7BD8
 
 | | Notum Fragment 872 | | Descriptor: | (2,5-dimethylphenyl) pyridine-4-carboxylate, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Zhao, Y, Jones, E.Y. | | Deposit date: | 2020-12-21 | | Release date: | 2022-01-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structural Analysis and Development of Notum Fragment Screening Hits.
Acs Chem Neurosci, 13, 2022
|
|
7BDF
 
 | | Notum Fragment 927 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-phenoxyethanoyl)piperazin-2-one, DIMETHYL SULFOXIDE, ... | | Authors: | Zhao, Y, Jones, E.Y. | | Deposit date: | 2020-12-21 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Analysis and Development of Notum Fragment Screening Hits.
Acs Chem Neurosci, 13, 2022
|
|
7BDH
 
 | | Notum Fragment 955 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, N-(2,1,3-Benzothiadiazol-5-yl)acetamide, ... | | Authors: | Zhao, Y, Jones, E.Y. | | Deposit date: | 2020-12-21 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structural Analysis and Development of Notum Fragment Screening Hits.
Acs Chem Neurosci, 13, 2022
|
|
7BCF
 
 | | Notum Fragment 722 | | Descriptor: | 2-[4-(2,5-Dioxopyrrolidin-1-yl)phenoxy]acetate, 2-acetamido-2-deoxy-beta-D-glucopyranose, Palmitoleoyl-protein carboxylesterase NOTUM, ... | | Authors: | Zhao, Y, Jones, E.Y. | | Deposit date: | 2020-12-19 | | Release date: | 2022-01-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural Analysis and Development of Notum Fragment Screening Hits.
Acs Chem Neurosci, 13, 2022
|
|
7B8M
 
 | | Notum-Fragment 193 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-methoxy-2-(1~{H}-pyrazol-3-yl)phenol, ... | | Authors: | Zhao, Y, Jonees, E.Y. | | Deposit date: | 2020-12-13 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural Analysis and Development of Notum Fragment Screening Hits.
Acs Chem Neurosci, 13, 2022
|
|
3CG7
 
 | |
4AWP
 
 | | Complex of HSP90 ATPase domain with tropane derived inhibitors | | Descriptor: | HEAT SHOCK PROTEIN HSP 90-ALPHA, N-{(3-endo)-8-[5-(benzylcarbamoyl)pyridin-2-yl]-8-azabicyclo[3.2.1]oct-3-yl}-2,5-dimethylbenzene-1,4-dicarboxamide | | Authors: | Lougheed, J.C, Stout, T.J. | | Deposit date: | 2012-06-05 | | Release date: | 2012-08-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Discovery of Xl888: A Novel Tropane-Derived Small Molecule Inhibitor of Hsp90.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
3CM6
 
 | |
7COW
 
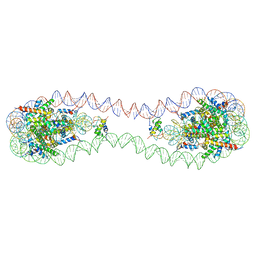 | | 353 bp di-nucleosome harboring cohesive DNA termini with linker histone H1.0 | | Descriptor: | CALCIUM ION, CHLORIDE ION, DNA (353-MER), ... | | Authors: | Adhireksan, Z, Sharma, D, Lee, P.L, Davey, C.A. | | Deposit date: | 2020-08-05 | | Release date: | 2021-08-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Engineering nucleosomes for generating diverse chromatin assemblies.
Nucleic Acids Res., 49, 2021
|
|
4AQ0
 
 | | Structure of the Gh92 Family Glycosyl Hydrolase Ccman5 in complex with deoxymannojirimycin | | Descriptor: | 1-DEOXYMANNOJIRIMYCIN, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Baranova, E, Tiels, P, Callewaert, N, Remaut, H. | | Deposit date: | 2012-04-12 | | Release date: | 2013-02-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | A Bacterial Glycosidase Enables Mannose-6-Phosphate Modification and Improved Cellular Uptake of Yeast-Produced Recombinant Human Lysosomal Enzymes.
Nat.Biotechnol., 30, 2012
|
|
2GOV
 
 | | Solution structure of Murine p22HBP | | Descriptor: | Heme-binding protein 1 | | Authors: | Volkman, B.F, Dias, J.S, Goodfellow, B.J, Peterson, F.C, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-04-14 | | Release date: | 2006-05-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The First Structure from the SOUL/HBP Family of Heme-binding Proteins, Murine P22HBP.
J.Biol.Chem., 281, 2006
|
|
3VO2
 
 | |
3VO1
 
 | |
3W3D
 
 | | Crystal structure of smooth muscle G actin DNase I complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, gamma-enteric smooth muscle, ... | | Authors: | Sakabe, N, Sakabe, K, Sasaki, K, Kondo, H, Shimomur, M. | | Deposit date: | 2012-12-20 | | Release date: | 2013-01-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Refined structure and solvent network of chicken gizzard G-actin DNase 1 complex at 1.8A resolution
Acta Crystallogr.,Sect.A, 49, 1993
|
|
