5D7D
 
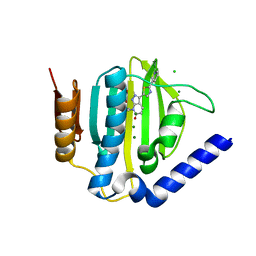 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a ligand | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 7-propyl-3-[2-(pyridin-3-yl)-1,3-thiazol-5-yl]-1,7-dihydro-6H-pyrazolo[3,4-b]pyridin-6-one, CHLORIDE ION, ... | | Authors: | Zhang, J, Yang, Q, Cross, J.B, Romero, J.A.C, Ryan, M.D, Lippa, B, Dolle, R.E, Andersen, O.A, Barker, J, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C. | | Deposit date: | 2015-08-13 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of Indazole Derivatives as a Novel Class of Bacterial Gyrase B Inhibitors.
Acs Med.Chem.Lett., 6, 2015
|
|
5D7R
 
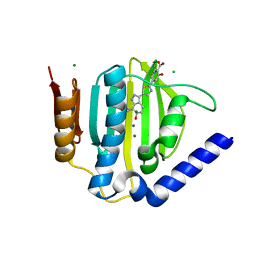 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a ligand | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-hydroxy-5-[5-(6-hydroxy-7-propyl-2H-indazol-3-yl)-1,3-thiazol-2-yl]pyridine-2-carboxylic acid, CHLORIDE ION, ... | | Authors: | Zhang, J, Yang, Q, Cross, J.B, Romero, J.A.C, Ryan, M.D, Lippa, B, Dolle, R.E, Andersen, O.A, Barker, J, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C. | | Deposit date: | 2015-08-14 | | Release date: | 2015-11-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery of Indazole Derivatives as a Novel Class of Bacterial Gyrase B Inhibitors.
Acs Med.Chem.Lett., 6, 2015
|
|
7T5H
 
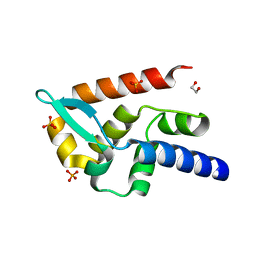 | | Structure of rabies virus phosphoprotein C-terminal domain, wild type | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, Phosphoprotein, ... | | Authors: | Zhan, J, Metcalfe, R.D, Gooley, P.R, Griffin, M.D.W. | | Deposit date: | 2021-12-12 | | Release date: | 2022-04-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular Basis of Functional Effects of Phosphorylation of the C-Terminal Domain of the Rabies Virus P Protein.
J.Virol., 96, 2022
|
|
7T5G
 
 | | Structure of rabies virus phosphoprotein C-terminal domain, S210E mutant | | Descriptor: | Phosphoprotein, SULFATE ION | | Authors: | Zhan, J, Metcalfe, R.D, Gooley, P.R, Griffin, M.D.W. | | Deposit date: | 2021-12-12 | | Release date: | 2022-04-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular Basis of Functional Effects of Phosphorylation of the C-Terminal Domain of the Rabies Virus P Protein.
J.Virol., 96, 2022
|
|
5JR9
 
 | | Crystal structure of apo-NeC3PO | | Descriptor: | NEQ131 | | Authors: | Zhang, J, Gan, J. | | Deposit date: | 2016-05-06 | | Release date: | 2016-09-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for single-stranded RNA recognition and cleavage by C3PO
Nucleic Acids Res., 44, 2016
|
|
5JRC
 
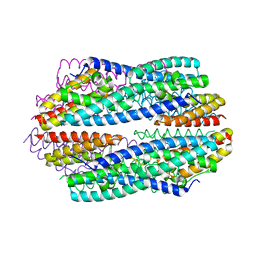 | |
4G42
 
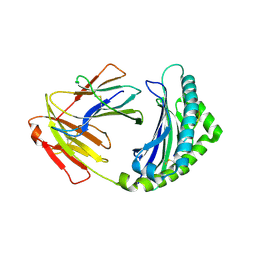 | | Structure of the Chicken MHC Class I Molecule BF2*0401 complexed to pepitde P8D | | Descriptor: | 8-MERIC PEPTIDE P8D, Beta-2 microglobulin, MHC class I alpha chain 2 | | Authors: | Zhang, J, Chen, Y, Qi, J, Gao, F, Liu, J, Kaufman, J, Xia, C, Gao, G.F. | | Deposit date: | 2012-07-16 | | Release date: | 2012-11-21 | | Method: | X-RAY DIFFRACTION (2.294 Å) | | Cite: | Narrow Groove and Restricted Anchors of MHC Class I Molecule BF2*0401 Plus Peptide Transporter Restriction Can Explain Disease Susceptibility of B4 Chickens.
J.Immunol., 189, 2012
|
|
4G43
 
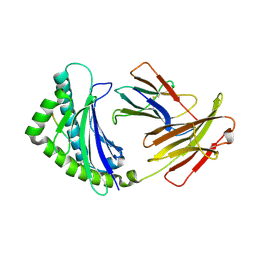 | | Structure of the chicken MHC class I molecule BF2*0401 complexed to P5E | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 8-MERIC PEPTIDE P5E, Beta-2 microglobulin, ... | | Authors: | Zhang, J, Chen, Y, Qi, J, Gao, F, Liu, J, Kaufman, J, Xia, C, Gao, G.F. | | Deposit date: | 2012-07-16 | | Release date: | 2012-11-21 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Narrow Groove and Restricted Anchors of MHC Class I Molecule BF2*0401 Plus Peptide Transporter Restriction Can Explain Disease Susceptibility of B4 Chickens.
J.Immunol., 189, 2012
|
|
3J03
 
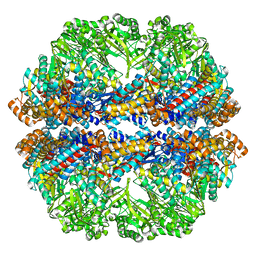 | | Lidless Mm-cpn in the closed state with ATP/AlFx | | Descriptor: | Lidless Mm-cpn | | Authors: | Zhang, J, Ma, B, DiMaio, F, Douglas, N.R, Joachimiak, L, Baker, D, Frydman, J, Levitt, M, Chiu, W. | | Deposit date: | 2011-02-10 | | Release date: | 2011-05-18 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Cryo-EM structure of a group II chaperonin in the prehydrolysis ATP-bound state leading to lid closure.
Structure, 19, 2011
|
|
7WNW
 
 | |
1F20
 
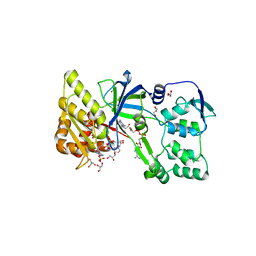 | | CRYSTAL STRUCTURE OF RAT NEURONAL NITRIC-OXIDE SYNTHASE FAD/NADP+ DOMAIN AT 1.9A RESOLUTION. | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, FORMIC ACID, GLYCEROL, ... | | Authors: | Zhang, J, Martasek, P, Masters, B.S, Kim, J.P. | | Deposit date: | 2000-05-22 | | Release date: | 2001-10-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the FAD/NADPH-binding domain of rat neuronal nitric-oxide synthase. Comparisons with NADPH-cytochrome P450 oxidoreductase.
J.Biol.Chem., 276, 2001
|
|
3IYF
 
 | | Atomic Model of the Lidless Mm-cpn in the Open State | | Descriptor: | Chaperonin | | Authors: | Zhang, J, Baker, M.L, Schroeder, G, Douglas, N.R, Reissmann, S, Jakana, J, Dougherty, M, Fu, C.J, Levitt, M, Ludtke, S.J, Frydman, J, Chiu, W. | | Deposit date: | 2009-10-23 | | Release date: | 2010-02-02 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Mechanism of folding chamber closure in a group II chaperonin
Nature, 463, 2010
|
|
4TZP
 
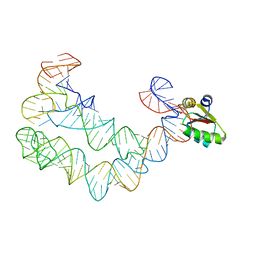 | | As Grown, Untreated Co-crystals of the Ternary Complex Containing a T-box Stem I RNA, its cognate tRNAGly, and B. subtilis YbxF protein | | Descriptor: | Ribosome-associated protein L7Ae-like, engineered tRNA, glyQ T-box Stem I | | Authors: | Zhang, J, Ferre-D'Amare, A.R. | | Deposit date: | 2014-07-10 | | Release date: | 2014-09-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (8.503 Å) | | Cite: | Dramatic Improvement of Crystals of Large RNAs by Cation Replacement and Dehydration.
Structure, 22, 2014
|
|
4TZV
 
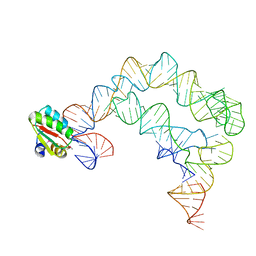 | | Co-crystals of the Ternary Complex Containing a T-box Stem I RNA, its Cognate tRNAGly, and B. subtilis YbxF protein, treated by removing lithium sulfate post crystallization | | Descriptor: | Ribosome-associated protein L7Ae-like, T-box stem I, engineered tRNA | | Authors: | Zhang, J, Ferre-D'Amare, A.R. | | Deposit date: | 2014-07-11 | | Release date: | 2014-09-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (5.03 Å) | | Cite: | Dramatic Improvement of Crystals of Large RNAs by Cation Replacement and Dehydration.
Structure, 22, 2014
|
|
4TZW
 
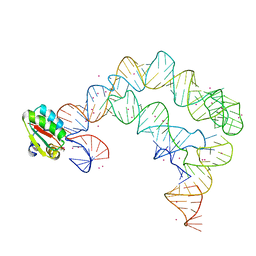 | | Co-crystals of the Ternary Complex Containing a T-box Stem I RNA, its Cognate tRNAGly, and B. subtilis YbxF protein, treated by removing lithium sulfate and replacing Mg2+ with Sr2+ post crystallization | | Descriptor: | Ribosome-associated protein L7Ae-like, STRONTIUM ION, T-box Stem I, ... | | Authors: | Zhang, J, Ferre-D'Amare, A.R. | | Deposit date: | 2014-07-11 | | Release date: | 2014-09-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (4.671 Å) | | Cite: | Dramatic Improvement of Crystals of Large RNAs by Cation Replacement and Dehydration.
Structure, 22, 2014
|
|
5I8I
 
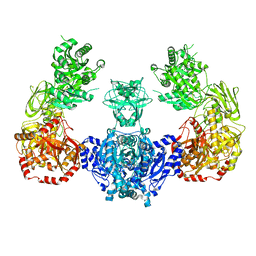 | |
3J9T
 
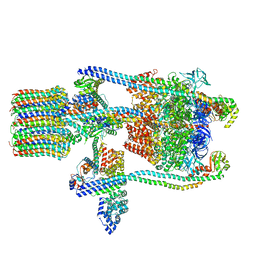 | | Yeast V-ATPase state 1 | | Descriptor: | V-type proton ATPase catalytic subunit A, V-type proton ATPase subunit B, V-type proton ATPase subunit C, ... | | Authors: | Zhao, J, Benlekbir, S, Rubinstein, J.L. | | Deposit date: | 2015-02-23 | | Release date: | 2015-05-13 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Electron cryomicroscopy observation of rotational states in a eukaryotic V-ATPase.
Nature, 521, 2015
|
|
3J9V
 
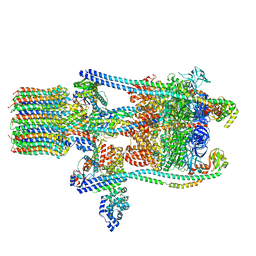 | | Yeast V-ATPase state 3 | | Descriptor: | V-type proton ATPase catalytic subunit A, V-type proton ATPase subunit B, V-type proton ATPase subunit C, ... | | Authors: | Zhao, J, Benlekbir, S, Rubinstein, J.L. | | Deposit date: | 2015-02-23 | | Release date: | 2015-05-13 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (8.3 Å) | | Cite: | Electron cryomicroscopy observation of rotational states in a eukaryotic V-ATPase.
Nature, 521, 2015
|
|
3J9U
 
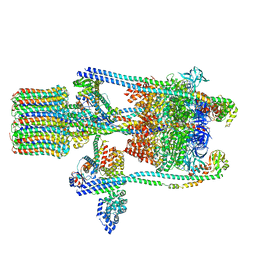 | | Yeast V-ATPase state 2 | | Descriptor: | V-type proton ATPase catalytic subunit A, V-type proton ATPase subunit B, V-type proton ATPase subunit C, ... | | Authors: | Zhao, J, Benlekbir, S, Rubinstein, J.L. | | Deposit date: | 2015-02-23 | | Release date: | 2015-05-13 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Electron cryomicroscopy observation of rotational states in a eukaryotic V-ATPase.
Nature, 521, 2015
|
|
2M8V
 
 | | Solution Structure and Activity Study of Bovicin HJ50, a Particular Type AII Lantibiotic | | Descriptor: | BovA | | Authors: | Zhang, J, Feng, Y, Wang, J, Zhong, J. | | Deposit date: | 2013-05-29 | | Release date: | 2014-05-21 | | Last modified: | 2024-07-10 | | Method: | SOLUTION NMR | | Cite: | Type AII lantibiotic bovicin HJ50 with a rare disulfide bond: structure, structure-activity relationships and mode of action.
Biochem.J., 461, 2014
|
|
3J02
 
 | | Lidless D386A Mm-cpn in the pre-hydrolysis ATP-bound state | | Descriptor: | Lidless D386A Mm-cpn variant | | Authors: | Zhang, J, Ma, B, DiMaio, F, Douglas, N.R, Joachimiak, L, Baker, D, Frydman, J, Levitt, M, Chiu, W. | | Deposit date: | 2011-02-10 | | Release date: | 2011-05-18 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Cryo-EM structure of a group II chaperonin in the prehydrolysis ATP-bound state leading to lid closure.
Structure, 19, 2011
|
|
4GZG
 
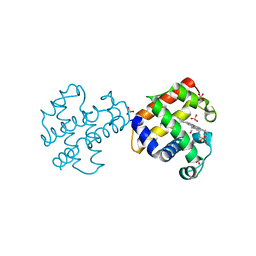 | | Crystal structures of DHPA-CO complex | | Descriptor: | CARBON MONOXIDE, Dehaloperoxidase A, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Zhao, J, Franzen, S. | | Deposit date: | 2012-09-06 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | The role of distal histidine in carbonmonoxide DHP structure
To be Published
|
|
8W8D
 
 | |
5VOX
 
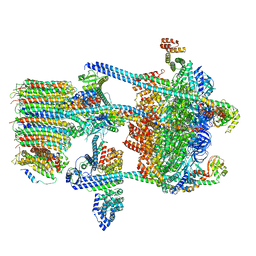 | | Yeast V-ATPase in complex with Legionella pneumophila effector SidK (rotational state 1) | | Descriptor: | V-type proton ATPase catalytic subunit A,V-type proton ATPase catalytic subunit A, V-type proton ATPase subunit B, V-type proton ATPase subunit C, ... | | Authors: | Zhao, J. | | Deposit date: | 2017-05-03 | | Release date: | 2017-06-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Molecular basis for the binding and modulation of V-ATPase by a bacterial effector protein.
PLoS Pathog., 13, 2017
|
|
3UIT
 
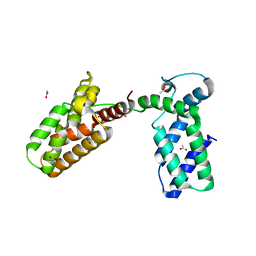 | | Overall structure of Patj/Pals1/Mals complex | | Descriptor: | ACETATE ION, InaD-like protein, MAGUK p55 subfamily member 5, ... | | Authors: | Zhang, J, Yang, X, Long, J, Shen, Y. | | Deposit date: | 2011-11-06 | | Release date: | 2012-02-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of an L27 domain heterotrimer from cell polarity complex Patj/Pals1/Mals2 reveals mutually independent L27 domain assembly mode
J.Biol.Chem., 287, 2012
|
|
