3W2A
 
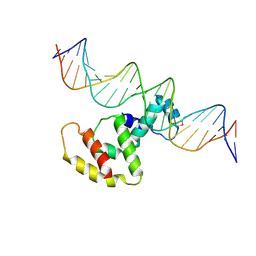 | | Crystal structure of VirB core domain complexed with the cis-acting site upstream icsp promoter | | Descriptor: | DNA (31-mer), Virulence regulon transcriptional activator VirB | | Authors: | Gao, X.P, Waltersperger, S, Wang, M.T, Cui, S. | | Deposit date: | 2012-11-27 | | Release date: | 2013-09-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.775 Å) | | Cite: | Structural insights into VirB-DNA complexes reveal mechanism of transcriptional activation of virulence genes
Nucleic Acids Res., 41, 2013
|
|
9IJL
 
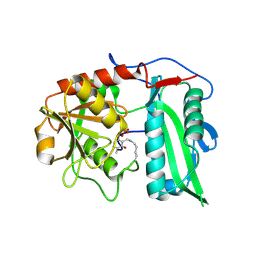 | |
9IVE
 
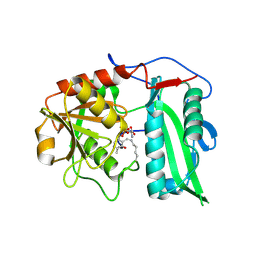 | | Structure of wild-type aminotransferase from Mycolicibacterium neoaurum in complex with LLP and ALA | | Descriptor: | ALANINE, Branched-chain amino acid transferase | | Authors: | Wei, H, Cong, L, You, S, Liu, W. | | Deposit date: | 2024-07-23 | | Release date: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structure-guided engineering an (R)-transaminase from Mycobacterium neoaurum for efficient synthesis of chiral N-heterocyclic amines.
Int.J.Biol.Macromol., 287, 2024
|
|
3OSY
 
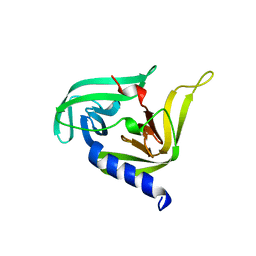 | |
5YBH
 
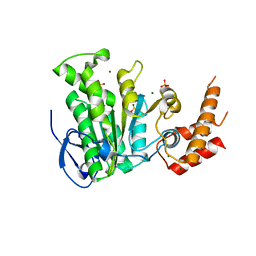 | |
5YBI
 
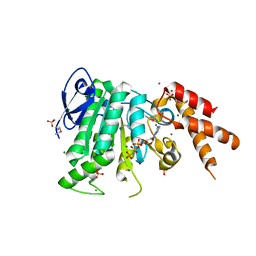 | | Structure of the bacterial pathogens ATPase with substrate AMPPNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Probable ATP synthase SpaL/MxiB, ... | | Authors: | Mu, Z.X, Gao, X.P, Cui, S. | | Deposit date: | 2017-09-05 | | Release date: | 2018-06-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.268 Å) | | Cite: | Structural Insight Into Conformational Changes Induced by ATP Binding in a Type III Secretion-Associated ATPase FromShigella flexneri.
Front Microbiol, 9, 2018
|
|
7YC2
 
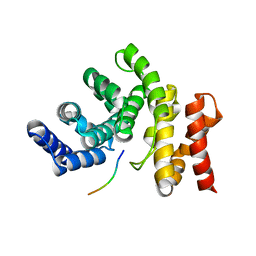 | |
5ZT1
 
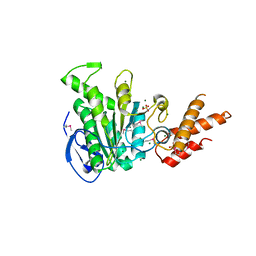 | | Structure of the bacterial pathogens ATPase with substrate ATP gamma S | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Probable ATP synthase SpaL/MxiB, ... | | Authors: | Gao, X.P, Mu, Z.X, Cui, S. | | Deposit date: | 2018-05-01 | | Release date: | 2018-05-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.114 Å) | | Cite: | Structural Insight Into Conformational Changes Induced by ATP Binding in a Type III Secretion-Associated ATPase FromShigella flexneri
Front Microbiol, 9, 2018
|
|
7XT3
 
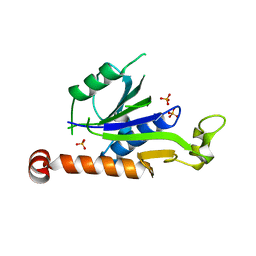 | | Crystal Structure of Hepatitis virus A 2C protein 128-335 aa | | Descriptor: | Genome polyprotein, PHOSPHATE ION | | Authors: | Chen, P, Wojdyla, J.A, Li, Z, Wang, M, Cui, S. | | Deposit date: | 2022-05-16 | | Release date: | 2022-07-27 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Biochemical and structural characterization of hepatitis A virus 2C reveals an unusual ribonuclease activity on single-stranded RNA.
Nucleic Acids Res., 50, 2022
|
|
5GNB
 
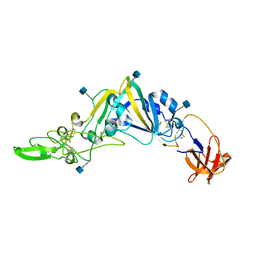 | | Crystal Structure of the Receptor Binding Domain of the Spike Glycoprotein of Human Betacoronavirus HKU1 (HKU1 1A-CTD, 2.3 angstrom, native-SAD phasing) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Guan, H, Wojdyla, J.A, Wang, M, Cui, S. | | Deposit date: | 2016-07-20 | | Release date: | 2017-06-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the receptor binding domain of the spike glycoprotein of human betacoronavirus HKU1
Nat Commun, 8, 2017
|
|
8YG1
 
 | | The Dimer Structure of DSR2 alone | | Descriptor: | SIR2-like domain-containing protein | | Authors: | Gao, X, Zhu, H, Cui, S. | | Deposit date: | 2024-02-26 | | Release date: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Activation of the bacterial defense-associated sirtuin system.
Commun Biol, 8, 2025
|
|
8YGA
 
 | | The tetramer Structure of DSR2 alone | | Descriptor: | SIR2-like domain-containing protein | | Authors: | Gao, X, Zhu, H, Cui, S. | | Deposit date: | 2024-02-26 | | Release date: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (4.53 Å) | | Cite: | Activation of the bacterial defense-associated sirtuin system.
Commun Biol, 8, 2025
|
|
8YGN
 
 | |
8YGO
 
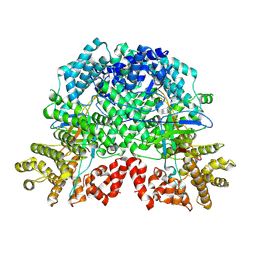 | | The complex by DSR2-CTD-SPR with NAD | | Descriptor: | SIR2-like domain-containing protein, SPR | | Authors: | Gao, X, Zhu, H, Cui, S. | | Deposit date: | 2024-02-26 | | Release date: | 2025-03-05 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Activation of the bacterial defense-associated sirtuin system.
Commun Biol, 8, 2025
|
|
8YGM
 
 | | The cryo-EM Structure of SPR | | Descriptor: | SPR | | Authors: | Gao, X, Zhu, H, Cui, S. | | Deposit date: | 2024-02-26 | | Release date: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | Activation of the bacterial defense-associated sirtuin system.
Commun Biol, 8, 2025
|
|
8YGP
 
 | |
8YGK
 
 | |
8YGC
 
 | | The Dimer Structure of DSR2-SPR | | Descriptor: | SIR2-like domain-containing protein, SPR | | Authors: | Gao, X, Zhu, H, Cui, S. | | Deposit date: | 2024-02-26 | | Release date: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (4.03 Å) | | Cite: | Activation of the bacterial defense-associated sirtuin system.
Commun Biol, 8, 2025
|
|
8YGF
 
 | |
8Y83
 
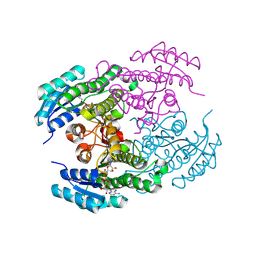 | | Crystal structure of a ketoreductase from Sphingobacterium siyangense SY1 with co-enzyme | | Descriptor: | NAD(P)-dependent dehydrogenase (Short-subunit alcohol dehydrogenase family), NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Zheng, Z.R, Wei, H.L, Liu, W.D, You, S. | | Deposit date: | 2024-02-05 | | Release date: | 2024-06-12 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based reshaping of a new ketoreductase from Sphingobacterium siyangense SY1 toward alpha-haloacetophenones.
Int.J.Biol.Macromol., 277, 2024
|
|
8Y7R
 
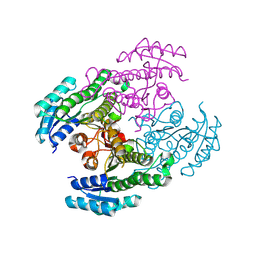 | |
8YOY
 
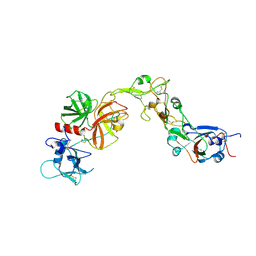 | | Structure of HKU1A RBD with TMPRSS2 | | Descriptor: | Spike protein S1, Transmembrane protease serine 2 | | Authors: | Gao, X, Cui, S, Ding, W, Shang, K, Zhu, H, Zhu, K. | | Deposit date: | 2024-03-14 | | Release date: | 2024-08-28 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structural basis for the interaction between human coronavirus HKU1 spike receptor binding domain and its receptor TMPRSS2.
Cell Discov, 10, 2024
|
|
8YQQ
 
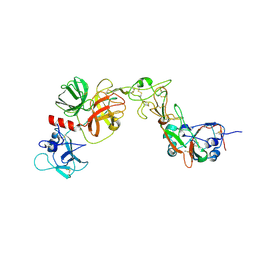 | | Structure of HKU1B RBD with TMPRSS2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, Transmembrane protease serine 2 | | Authors: | Gao, X, Cui, S, Ding, W, Zhu, K, Shang, K, Zhu, H. | | Deposit date: | 2024-03-19 | | Release date: | 2024-08-28 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.95 Å) | | Cite: | Structural basis for the interaction between human coronavirus HKU1 spike receptor binding domain and its receptor TMPRSS2.
Cell Discov, 10, 2024
|
|
8K88
 
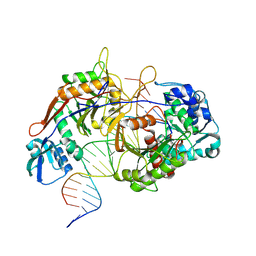 | | Structure of procaryotic Ago | | Descriptor: | DNA (41-mer), DNA/RNA (21-mer), MAGNESIUM ION, ... | | Authors: | Gao, X, Sun, D, Cui, S, Wang, Y. | | Deposit date: | 2023-07-29 | | Release date: | 2024-07-03 | | Last modified: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Nucleic acid-induced NADase activation of a short Sir2-associated prokaryotic Argonaute system.
Cell Rep, 43, 2024
|
|
8K87
 
 | | Dimer structure of procaryotic Ago | | Descriptor: | DNA (41-mer), MAGNESIUM ION, Piwi domain protein, ... | | Authors: | Gao, X, Sun, D, Cui, S, Wang, Y. | | Deposit date: | 2023-07-29 | | Release date: | 2024-07-03 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Nucleic acid-induced NADase activation of a short Sir2-associated prokaryotic Argonaute system.
Cell Rep, 43, 2024
|
|
