8PNO
 
 | |
5HSA
 
 | | Alcohol Oxidase AOX1 from Pichia Pastoris | | Descriptor: | ARABINO-FLAVIN-ADENINE DINUCLEOTIDE, Alcohol oxidase 1, CALCIUM ION, ... | | Authors: | Neumann, P, Ficner, R, Feussner, I, Koch, C. | | Deposit date: | 2016-01-25 | | Release date: | 2016-03-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of Alcohol Oxidase from Pichia pastoris.
Plos One, 11, 2016
|
|
3OCI
 
 | | Crystal structure of TBP (TATA box binding protein) | | Descriptor: | 1,2-ETHANEDIOL, TRANSCRIPTION INITIATION FACTOR TFIID (TFIID-1) | | Authors: | Cui, S, Wollmann, P, Moldt, M, Hopfner, K.-P. | | Deposit date: | 2010-08-10 | | Release date: | 2011-07-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Structure and mechanism of the Swi2/Snf2 remodeller Mot1 in complex with its substrate TBP.
Nature, 475, 2011
|
|
5L4W
 
 | | Crystal structure of FimH lectin domain in complex with 3-Fluoro-Heptylmannoside | | Descriptor: | Protein FimH, heptyl 3-fluoro-alpha-D-mannopyranoside | | Authors: | Jakob, R.P, Zihlmann, P, Rabbani, S, Maier, T, Ernst, B. | | Deposit date: | 2016-05-26 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-Affinity Carbohydrate-Lectin Interactions: How Nature Makes it Possible
To Be Published
|
|
5L4X
 
 | | Crystal structure of FimH lectin domain in complex with 4-Deoxy-Heptylmannoside | | Descriptor: | Protein FimH, heptyl 4-deoxy-4-deoxy-alpha-D-mannopyranoside | | Authors: | Jakob, R.P, Zihlmann, P, Rabbani, S, Maier, T, Ernst, B. | | Deposit date: | 2016-05-26 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-Affinity Carbohydrate-Lectin Interactions: How Nature Makes it Possible
To Be Published
|
|
3MWQ
 
 | |
3MWR
 
 | |
4KXX
 
 | | Human transketolase in covalent complex with donor ketose D-sedoheptulose-7-phosphate | | Descriptor: | (2R,3R,4S,5R,6S)-2,3,4,5,6,7-hexahydroxyheptyl dihydrogen phosphate, 1,2-ETHANEDIOL, MAGNESIUM ION, ... | | Authors: | Neumann, P, Luedtke, S, Ficner, R, Tittmann, K. | | Deposit date: | 2013-05-28 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Sub-angstrom-resolution crystallography reveals physical distortions that enhance reactivity of a covalent enzymatic intermediate.
Nat Chem, 5, 2013
|
|
4KXU
 
 | | Human transketolase in covalent complex with donor ketose D-fructose-6-phosphate | | Descriptor: | 1,2-ETHANEDIOL, D-SORBITOL-6-PHOSPHATE, MAGNESIUM ION, ... | | Authors: | Neumann, P, Luedtke, S, Ficner, R, Tittmann, K. | | Deposit date: | 2013-05-28 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Sub-angstrom-resolution crystallography reveals physical distortions that enhance reactivity of a covalent enzymatic intermediate.
Nat Chem, 5, 2013
|
|
4KXY
 
 | | Human transketolase in complex with ThDP analogue (R)-2-(1,2-dihydroxyethyl)-3-deaza-ThDP | | Descriptor: | 1,2-ETHANEDIOL, 2-{4-[(4-amino-2-methylpyrimidin-5-yl)methyl]-5-[(1R)-1,2-dihydroxyethyl]-3-methylthiophen-2-yl}ethyl trihydrogen diphosphate, CALCIUM ION, ... | | Authors: | Neumann, P, Luedtke, S, Erixon, K.M, Leeper, F, Kluger, R, Ficner, R, Tittmann, K. | | Deposit date: | 2013-05-28 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Sub-angstrom-resolution crystallography reveals physical distortions that enhance reactivity of a covalent enzymatic intermediate.
Nat Chem, 5, 2013
|
|
4KXW
 
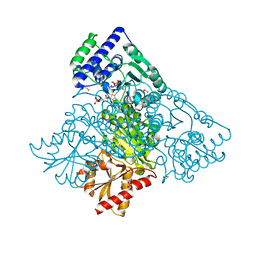 | | Human transketolase in covalent complex with donor ketose D-xylulose-5-phosphate, crystal 2 | | Descriptor: | 1,2-ETHANEDIOL, D-XYLITOL-5-PHOSPHATE, MAGNESIUM ION, ... | | Authors: | Neumann, P, Luedtke, S, Ficner, R, Tittmann, K. | | Deposit date: | 2013-05-28 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Sub-angstrom-resolution crystallography reveals physical distortions that enhance reactivity of a covalent enzymatic intermediate.
Nat Chem, 5, 2013
|
|
4KXV
 
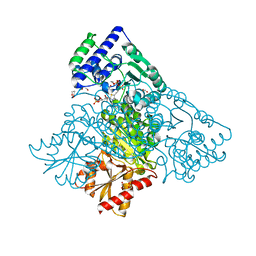 | | Human transketolase in covalent complex with donor ketose D-xylulose-5-phosphate, crystal 1 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, D-XYLITOL-5-PHOSPHATE, ... | | Authors: | Neumann, P, Luedtke, S, Ficner, R, Tittmann, K. | | Deposit date: | 2013-05-28 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Sub-angstrom-resolution crystallography reveals physical distortions that enhance reactivity of a covalent enzymatic intermediate.
Nat Chem, 5, 2013
|
|
3FT7
 
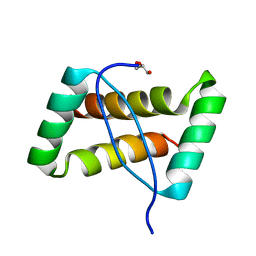 | | Crystal structure of an extremely stable dimeric protein from sulfolobus islandicus | | Descriptor: | GLYCEROL, Uncharacterized protein ORF56 | | Authors: | Neumann, P, Loew, C, Weininger, U, Stubbs, M.T. | | Deposit date: | 2009-01-12 | | Release date: | 2009-10-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Stability Analysis of an Extremely Stable Dimeric DNA Binding Protein from Sulfolobus islandicus
Biochemistry, 48, 2009
|
|
4KGD
 
 | |
8ARU
 
 | |
3FD7
 
 | | Crystal structure of Onconase C87A/C104A-ONC | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Protein P-30, ... | | Authors: | Neumann, P, Schulenburg, C, Arnold, U, Ulbrich-Hofmann, R, Stubbs, M.T. | | Deposit date: | 2008-11-25 | | Release date: | 2009-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.531 Å) | | Cite: | Impact of the C-terminal disulfide bond on the folding and stability of onconase.
Chembiochem, 11, 2010
|
|
3CGN
 
 | | Crystal Structure of thermophilic SlyD | | Descriptor: | Peptidyl-prolyl cis-trans isomerase, SULFATE ION | | Authors: | Neumann, P, Loew, C, Stubbs, M.T, Balbach, J. | | Deposit date: | 2008-03-06 | | Release date: | 2009-03-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure Determination and Functional Characterization of the Metallochaperone SlyD from Thermus thermophilus
J.Mol.Biol., 398, 2010
|
|
5L4U
 
 | | Crystal structure of FimH lectin domain in complex with 2-Fluoro-Heptylmannoside | | Descriptor: | Protein FimH, heptyl 2-fluoro-alpha-D-mannopyranoside | | Authors: | Jakob, R.P, Zihlmann, P, Rabbani, S, Maier, T, Ernst, B. | | Deposit date: | 2016-05-26 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High-Affinity Carbohydrate-Lectin Interactions: How Nature Makes it Possible
To Be Published
|
|
5L4Y
 
 | | Crystal structure of FimH lectin domain in complex with 4-Fluoro-Heptylmannoside | | Descriptor: | Protein FimH, heptyl 4-deoxy-4-fluoro-alpha-D-mannopyranoside | | Authors: | Jakob, R.P, Zihlmann, P, Rabbani, S, Maier, T, Ernst, B. | | Deposit date: | 2016-05-26 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-Affinity Carbohydrate-Lectin Interactions: How Nature Makes it Possible
To Be Published
|
|
5L4V
 
 | | Crystal structure of FimH lectin domain in complex with 3-Deoxy-Heptylmannoside | | Descriptor: | Protein FimH, heptyl 3-deoxy-alpha-D-mannopyranoside | | Authors: | Jakob, R.P, Zihlmann, P, Rabbani, S, Maier, T, Ernst, T. | | Deposit date: | 2016-05-26 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | High-Affinity Carbohydrate-Lectin Interactions: How Nature Makes it Possible
To Be Published
|
|
5L4T
 
 | | Crystal structure of FimH lectin domain in complex with 2-Deoxy-Heptylmannoside | | Descriptor: | Protein FimH, heptyl 2-deoxy-alpha-D-mannopyranoside | | Authors: | Jakob, R.P, Zihlmann, P, Rabbani, S, Maier, T, Ernst, B. | | Deposit date: | 2016-05-26 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-Affinity Carbohydrate-Lectin Interactions: How Nature Makes it Possible
To Be Published
|
|
5NHX
 
 | | Periplasmic domain of Outer Membrane Protein A from Klebsiella pneumoniae | | Descriptor: | CITRIC ACID, Outer membrane protein A | | Authors: | Demange, P, Tranier, S, Nars, G, Iordanov, I, Mourey, L, Saurel, O, Milon, A. | | Deposit date: | 2017-03-22 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and dynamics of the C-terminal domain of OmpA from Klebsiella pneumonia
To Be Published
|
|
4NHO
 
 | | Structure of the spliceosomal DEAD-box protein Prp28 | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, GLYCEROL, MERCURY (II) ION, ... | | Authors: | Moehlmann, S, Neumann, P, Ficner, R. | | Deposit date: | 2013-11-05 | | Release date: | 2014-06-18 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional analysis of the human spliceosomal DEAD-box helicase Prp28.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4EX7
 
 | | Crystal structure of the alnumycin P phosphatase in complex with free phosphate | | Descriptor: | AlnB, BORIC ACID, MAGNESIUM ION, ... | | Authors: | Oja, T, Niiranen, L, Sandalova, T, Klika, K.D, Niemi, J, Mantsala, P, Schneider, G, Metsa-Ketela, M. | | Deposit date: | 2012-04-30 | | Release date: | 2013-01-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for C-ribosylation in the alnumycin A biosynthetic pathway.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4EX8
 
 | | Crystal structure of the prealnumycin C-glycosynthase AlnA | | Descriptor: | AlnA, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Oja, T, Niiranen, L, Sandalova, T, Klika, K.D, Niemi, J, Mantsala, P, Schneider, G, Metsa-Ketela, M. | | Deposit date: | 2012-04-30 | | Release date: | 2013-01-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for C-ribosylation in the alnumycin A biosynthetic pathway.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
