8B2R
 
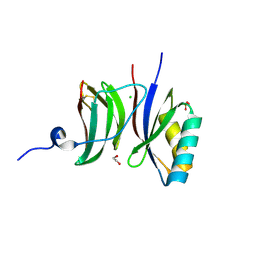 | |
1E3S
 
 | | Rat brain 3-hydroxyacyl-CoA dehydrogenase binary complex with NADH | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SHORT CHAIN 3-HYDROXYACYL-COA DEHYDROGENASE | | Authors: | Powell, A.J, Read, J.A, Banfield, M.J, Brady, R.L. | | Deposit date: | 2000-06-22 | | Release date: | 2001-05-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Recognition of Structurally Diverse Substrates by Type II 3-Hydroxyacyl-Coa Dehydrogenase (Hadh II) Amyloid-Beta Binding Alcohol Dehydrogenase (Abad)
J.Mol.Biol., 303, 2000
|
|
2JKW
 
 | | Pseudoazurin M16F | | Descriptor: | COPPER (II) ION, PSEUDOAZURIN | | Authors: | Yanagisawa, S, Crowley, P.B, Firbank, S.J, Lawler, A.T, Hunter, D.M, McFarlane, W, Li, C, Kohzuma, T, Banfield, M.J, Dennison, C. | | Deposit date: | 2008-09-01 | | Release date: | 2008-11-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Pi-Interaction Tuning of the Active Site Properties of Metalloproteins.
J.Am.Chem.Soc., 130, 2008
|
|
2VQA
 
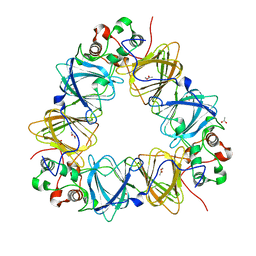 | | Protein-folding location can regulate Mn versus Cu- or Zn-binding. Crystal Structure of MncA. | | Descriptor: | ACETATE ION, MANGANESE (II) ION, SLL1358 PROTEIN | | Authors: | Tottey, S, Waldron, K.J, Firbank, S.J, Reale, B, Bessant, C, Sato, K, Gray, J, Banfield, M.J, Dennison, C, Robinson, N.J. | | Deposit date: | 2008-03-12 | | Release date: | 2008-10-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Protein-Folding Location Can Regulate Manganese-Binding Versus Copper- or Zinc-Binding.
Nature, 455, 2008
|
|
2VN3
 
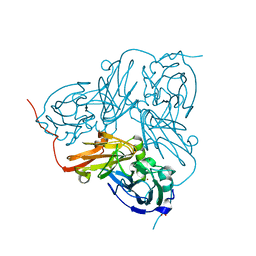 | | Nitrite Reductase from Alcaligenes xylosoxidans | | Descriptor: | COPPER (II) ION, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE, SULFATE ION, ... | | Authors: | Sato, K, Firbank, S.J, Li, C, Banfield, M.J, Dennison, C. | | Deposit date: | 2008-01-30 | | Release date: | 2008-12-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The Importance of the Long Type 1 Copper-Binding Loop of Nitrite Reductase for Structure and Function.
Chemistry, 14, 2008
|
|
2XI9
 
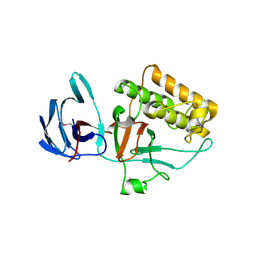 | | Pilus-presented adhesin, Spy0125 (Cpa), P1 form | | Descriptor: | ANCILLARY PROTEIN 1 | | Authors: | Pointon, J.A, Smith, W.D, Saalbach, G, Crow, A, Kehoe, M.A, Banfield, M.J. | | Deposit date: | 2010-06-28 | | Release date: | 2010-08-04 | | Last modified: | 2019-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Highly Unusual Thioester Bond in a Pilus Adhesin is Required for Efficient Host Cell Interaction
J.Biol.Chem., 285, 2010
|
|
2XMK
 
 | | Visualising the Metal-binding Versatility of Copper Trafficking Sites: Atx1 side-to-side (anaerobic) | | Descriptor: | CHLORIDE ION, COPPER (II) ION, SODIUM ION, ... | | Authors: | Badarau, A, Firbank, S.J, McCarthy, A.A, Banfield, M.J, Dennison, C. | | Deposit date: | 2010-07-28 | | Release date: | 2010-08-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Visualizing the Metal-Binding Versatility of Copper Trafficking Sites .
Biochemistry, 49, 2010
|
|
2XIC
 
 | | Pilus-presented adhesin, Spy0125 (Cpa), P212121 form (ESRF data) | | Descriptor: | ANCILLARY PROTEIN 1 | | Authors: | Pointon, J.A, Smith, W.D, Saalbach, G, Crow, A, Kehoe, M.A, Banfield, M.J. | | Deposit date: | 2010-06-28 | | Release date: | 2010-08-04 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A Highly Unusual Thioester Bond in a Pilus Adhesin is Required for Efficient Host Cell Interaction
J.Biol.Chem., 285, 2010
|
|
2XMV
 
 | | Copper chaperone Atx1 from Synechocystis PCC6803 (Cu1, trimeric form, His61Tyr mutant) | | Descriptor: | COPPER (I) ION, SSR2857 PROTEIN | | Authors: | Badarau, A, Firbank, S.J, McCarthy, A.A, Banfield, M.J, Dennison, C. | | Deposit date: | 2010-07-29 | | Release date: | 2010-08-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Visualizing the Metal-Binding Versatility of Copper Trafficking Sites .
Biochemistry, 49, 2010
|
|
2XV3
 
 | | Pseudomonas aeruginosa Azurin with mutated metal-binding loop sequence (CAAAAHAAAAM), chemically reduced, pH5.3 | | Descriptor: | AZURIN, COPPER (I) ION | | Authors: | Li, C, Sato, K, Monari, S, Salard, I, Sola, M, Banfield, M.J, Dennison, C. | | Deposit date: | 2010-10-22 | | Release date: | 2010-12-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Metal-Binding Loop Length is a Determinant of the Pka of a Histidine Ligand at a Type 1 Copper Site
Inorg.Chem., 50, 2011
|
|
2XID
 
 | | Pilus-presented adhesin, Spy0125 (Cpa), P212121 form (DLS) | | Descriptor: | ANCILLARY PROTEIN 1 | | Authors: | Pointon, J.A, Smith, W.D, Saalbach, G, Crow, A, Kehoe, M.A, Banfield, M.J. | | Deposit date: | 2010-06-28 | | Release date: | 2010-08-04 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | A Highly Unusual Thioester Bond in a Pilus Adhesin Required for Efficient Host Cell Interaction
J.Biol.Chem., 285, 2010
|
|
2XMJ
 
 | | Visualising the Metal-binding Versatility of Copper Trafficking Sites: Atx1 side-to-side (aerobic) | | Descriptor: | CHLORIDE ION, COPPER (II) ION, SODIUM ION, ... | | Authors: | Badarau, A, Firbank, S.J, McCarthy, A.A, Banfield, M.J, Dennison, C. | | Deposit date: | 2010-07-28 | | Release date: | 2010-08-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Visualizing the Metal-Binding Versatility of Copper Trafficking Sites .
Biochemistry, 49, 2010
|
|
2XMW
 
 | | PacS, N-terminal domain, from Synechocystis PCC6803 | | Descriptor: | CATION-TRANSPORTING ATPASE PACS, COPPER (I) ION | | Authors: | Badarau, A, Firbank, S.J, McCarthy, A.A, Banfield, M.J, Dennison, C. | | Deposit date: | 2010-07-29 | | Release date: | 2010-08-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Visualizing the Metal-Binding Versatility of Copper Trafficking Sites .
Biochemistry, 49, 2010
|
|
2VMJ
 
 | | Type 1 Copper-Binding Loop of Nitrite Reductase mutant: 130- CAPEGMVPWHVVSGM-144 to 130-CTPHPFM-136 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE, ZINC ION | | Authors: | Sato, K, Firbank, S.J, Li, C, Banfield, M.J, Dennison, C. | | Deposit date: | 2008-01-28 | | Release date: | 2008-12-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Importance of the Long Type 1 Copper-Binding Loop of Nitrite Reductase for Structure and Function.
Chemistry, 14, 2008
|
|
2XMT
 
 | | Copper chaperone Atx1 from Synechocystis PCC6803 (Cu1 form) | | Descriptor: | COPPER (I) ION, SSR2857 PROTEIN | | Authors: | Badarau, A, Firbank, S.J, McCarthy, A.A, Banfield, M.J, Dennison, C. | | Deposit date: | 2010-07-29 | | Release date: | 2010-08-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Visualizing the Metal-Binding Versatility of Copper Trafficking Sites .
Biochemistry, 49, 2010
|
|
2XV0
 
 | | Pseudomonas aeruginosa Azurin with mutated metal-binding loop sequence (CAAHAAM), chemically reduced, pH4.8 | | Descriptor: | AZURIN, COPPER (I) ION | | Authors: | Li, C, Sato, K, Monari, S, Salard, I, Sola, M, Banfield, M.J, Dennison, C. | | Deposit date: | 2010-10-22 | | Release date: | 2010-12-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Metal-Binding Loop Length is a Determinant of the Pka of a Histidine Ligand at a Type 1 Copper Site
Inorg.Chem., 50, 2011
|
|
2XV2
 
 | | Pseudomonas aeruginosa Azurin with mutated metal-binding loop sequence (CAAHAAM), chemically reduced, pH4.2 | | Descriptor: | AZURIN, COPPER (I) ION | | Authors: | Li, C, Sato, K, Monari, S, Salard, I, Sola, M, Banfield, M.J, Dennison, C. | | Deposit date: | 2010-10-22 | | Release date: | 2010-12-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Metal-Binding Loop Length is a Determinant of the Pka of a Histidine Ligand at a Type 1 Copper Site
Inorg.Chem., 50, 2011
|
|
2XMU
 
 | | Copper chaperone Atx1 from Synechocystis PCC6803 (Cu2 form) | | Descriptor: | COPPER (I) ION, SSR2857 PROTEIN | | Authors: | Badarau, A, Firbank, S.J, McCarthy, A.A, Banfield, M.J, Dennison, C. | | Deposit date: | 2010-07-29 | | Release date: | 2010-08-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Visualizing the Metal-Binding Versatility of Copper Trafficking Sites .
Biochemistry, 49, 2010
|
|
2XMM
 
 | | Visualising the Metal-binding Versatility of Copper Trafficking Sites: H61Y Atx1 side-to-side | | Descriptor: | CHLORIDE ION, COPPER (II) ION, SODIUM ION, ... | | Authors: | Badarau, A, Firbank, S.J, McCarthy, A.A, Banfield, M.J, Dennison, C. | | Deposit date: | 2010-07-28 | | Release date: | 2010-08-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Visualizing the Metal-Binding Versatility of Copper Trafficking Sites .
Biochemistry, 49, 2010
|
|
2VOZ
 
 | | Apo FutA2 from Synechocystis PCC6803 | | Descriptor: | PERIPLASMIC IRON-BINDING PROTEIN, SULFATE ION | | Authors: | Badarau, A, Firbank, S.J, Banfield, M.J, Dennison, C. | | Deposit date: | 2008-02-25 | | Release date: | 2008-03-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Futa2 is a Ferric Binding Protein from Synechocystis Pcc 6803.
J.Biol.Chem., 283, 2008
|
|
2VP1
 
 | | Fe-FutA2 from Synechocystis PCC6803 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, FE (III) ION, ... | | Authors: | Badarau, A, Firbank, S.J, Banfield, M.J, Dennison, C. | | Deposit date: | 2008-02-26 | | Release date: | 2008-03-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Futa2 is a Ferric Binding Protein from Synechocystis Pcc 6803.
J.Biol.Chem., 283, 2008
|
|
6FUD
 
 | |
6FUB
 
 | |
6G11
 
 | |
6GU1
 
 | |
