6ZAF
 
 | | Room temperature XFEL Isopenicillin N synthase structure in complex with Fe and ACV after exposure to dioxygen for 400ms. | | Descriptor: | FE (II) ION, Isopenicillin N synthase, L-D-(A-AMINOADIPOYL)-L-CYSTEINYL-D-VALINE, ... | | Authors: | Rabe, P, Kamps, J.J.A.G, Sutherlin, K, Pharm, C, McDonough, M.A, Leissing, T.M, Aller, P, Butryn, A, Linyard, J, Lang, P, Brem, J, Fuller, F.D, Batyuk, A, Hunter, M.S, Pettinati, I, Clifton, I.J, Alonso-Mori, R, Gul, S, Young, I, Kim, I, Bhowmick, A, ORiordan, L, Brewster, A.S, Claridge, T.D.W, Sauter, N.K, Yachandra, V, Yano, J, Kern, J.F, Orville, A.M, Schofield, C.J. | | Deposit date: | 2020-06-05 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4001 Å) | | Cite: | X-ray free-electron laser studies reveal correlated motion during isopenicillin N synthase catalysis.
Sci Adv, 7, 2021
|
|
6ZAJ
 
 | | Room temperature XFEL Isopenicillin N synthase structure in complex with Fe, O2 and ACV after exposure to dioxygen for 3000ms. | | Descriptor: | FE (II) ION, Isopenicillin N synthase, L-D-(A-AMINOADIPOYL)-L-CYSTEINYL-D-VALINE, ... | | Authors: | Rabe, P, Kamps, J.J.A.G, Sutherlin, K, Pharm, C, McDonough, M.A, Leissing, T.M, Aller, P, Butryn, A, Linyard, J, Lang, P, Brem, J, Fuller, F.D, Batyuk, A, Hunter, M.S, Pettinati, I, Clifton, I.J, Alonso-Mori, R, Gul, S, Young, I, Kim, I, Bhowmick, A, ORiordan, L, Brewster, A.S, Claridge, T.D.W, Sauter, N.K, Yachandra, V, Yano, J, Kern, J.F, Orville, A.M, Schofield, C.J. | | Deposit date: | 2020-06-05 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5301 Å) | | Cite: | X-ray free-electron laser studies reveal correlated motion during isopenicillin N synthase catalysis.
Sci Adv, 7, 2021
|
|
6ZAM
 
 | | Fluorine labeled IPNS S55C in complex with Fe and ACV under anaerobic conditions. | | Descriptor: | 1,1,1-tris(fluoranyl)propan-2-one, 1,1,1-tris(fluoranyl)propane-2,2-diol, FE (III) ION, ... | | Authors: | Rabe, P, Kamps, J.J.A.G, Sutherlin, K, Pharm, C, McDonough, M.A, Leissing, T.M, Aller, P, Butryn, A, Linyard, J, Lang, P, Brem, J, Fuller, F.D, Batyuk, A, Hunter, M.S, Pettinati, I, Clifton, I.J, Alonso-Mori, R, Gul, S, Young, I, Kim, I, Bhowmick, A, ORiordan, L, Brewster, A.S, Claridge, T.D.W, Sauter, N.K, Yachandra, V, Yano, J, Kern, J.F, Orville, A.M, Schofield, C.J. | | Deposit date: | 2020-06-05 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | X-ray free-electron laser studies reveal correlated motion during isopenicillin N synthase catalysis.
Sci Adv, 7, 2021
|
|
4BTB
 
 | |
6ZW8
 
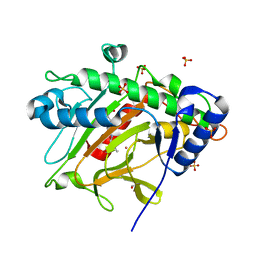 | | Isopenicillin N synthase in complex with Cd and ACV. | | Descriptor: | CADMIUM ION, GLYCEROL, Isopenicillin N synthase, ... | | Authors: | Rabe, P, Kamps, J.J.A.G, Sutherlin, K, Pharm, C, McDonough, M.A, Leissing, T.M, Aller, P, Butryn, A, Linyard, J, Lang, P, Brem, J, Fuller, F.D, Batyuk, A, Hunter, M.S, Pettinati, I, Clifton, I.J, Alonso-Mori, R, Gul, S, Young, I, Kim, I, Bhowmick, A, ORiordan, L, Brewster, A.S, Claridge, T.D.W, Sauter, N.K, Yachandra, V, Yano, J, Kern, J.F, Orville, A.M, Schofield, C.J. | | Deposit date: | 2020-07-27 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | X-ray free-electron laser studies reveal correlated motion during isopenicillin N synthase catalysis.
Sci Adv, 7, 2021
|
|
6Z5O
 
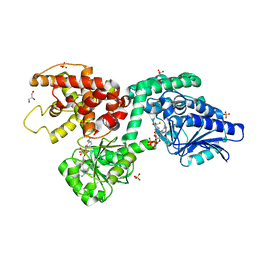 | | CRYSTAL STRUCTURE OF RAT PEROXISOMAL MULTIFUNCTIONAL ENZYME TYPE-1 (RPMFE1) COMPLEXED WITH COENZYME-A AND OXIDISED NICOTINAMIDE ADENINE DINUCLEOTIDE | | Descriptor: | COENZYME A, GLYCEROL, NICOTINAMIDE, ... | | Authors: | Wierenga, R.K, Sridhar, S, Kiema, T.R. | | Deposit date: | 2020-05-27 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystallographic binding studies of rat peroxisomal multifunctional enzyme type 1 with 3-ketodecanoyl-CoA: capturing active and inactive states of its hydratase and dehydrogenase catalytic sites.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
5AB4
 
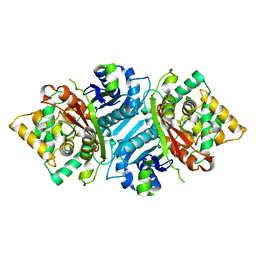 | |
5AB7
 
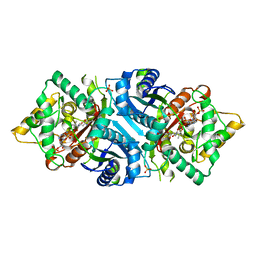 | |
5AB5
 
 | |
4BTA
 
 | | CRYSTAL STRUCTURE OF THE PEPTIDE(PRO-PRO-GLY)3 BOUND COMPLEX OF N- TERMINAL DOMAIN AND PEPTIDE SUBSTRATE BINDING DOMAIN OF PROLYL-4 HYDROXYLASE (RESIDUES 1-244) TYPE I FROM HUMAN | | Descriptor: | PROLINE RICH PEPTIDE, PROLYL 4-HYDROXYLASE SUBUNIT ALPHA-1 | | Authors: | Anantharajan, J, Koski, M.K, Pekkala, M, Wierenga, R.K. | | Deposit date: | 2013-06-14 | | Release date: | 2013-10-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | The Structural Motifs for Substrate Binding and Dimerization of the Alpha Subunit of Collagen Prolyl 4-Hydroxylase
Structure, 21, 2013
|
|
5AB6
 
 | |
4BT9
 
 | |
4BT8
 
 | |
3BA0
 
 | | Crystal structure of full-length human MMP-12 | | Descriptor: | ACETOHYDROXAMIC ACID, CALCIUM ION, Macrophage metalloelastase, ... | | Authors: | Bertini, I, Calderone, V, Fragai, M, Jaiswal, R, Luchinat, C, Melikian, M, Myonas, E, Svergun, D.I. | | Deposit date: | 2007-11-07 | | Release date: | 2008-07-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Evidence of reciprocal reorientation of the catalytic and hemopexin-like domains of full-length MMP-12.
J.Am.Chem.Soc., 130, 2008
|
|
5MG0
 
 | | Structure of PAS-GAF fragment of Deinococcus phytochrome by serial femtosecond crystallography | | Descriptor: | 1,2-ETHANEDIOL, 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, Bacteriophytochrome, ... | | Authors: | Burgie, E.S, Fuller, F.D, Gul, S, Miller, M.D, Young, I.D, Brewster, A.S, Clinger, J, Aller, P, Braeuer, P, Hutchison, C, Alonso-Mori, R, Kern, J, Yachandra, V.K, Yano, J, Sauter, N.K, Phillips Jr, G.N, Vierstra, R.D, Orville, A.M. | | Deposit date: | 2016-11-20 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Drop-on-demand sample delivery for studying biocatalysts in action at X-ray free-electron lasers.
Nat. Methods, 14, 2017
|
|
5MG1
 
 | | Structure of the photosensory module of Deinococcus phytochrome by serial femtosecond X-ray crystallography | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, Bacteriophytochrome | | Authors: | Burgie, E.S, Fuller, F.D, Gul, S, Young, I.D, Brewster, A.S, Clinger, J, Andi, B, Stan, C, Allaire, M, Nelsen, S, Alonso-Mori, R, Phillips Jr, G.N, Sauter, N.K, Kern, J, Yachandra, V.K, Yano, J, Vierstra, R.D, Orville, A.M. | | Deposit date: | 2016-11-20 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Drop-on-demand sample delivery for studying biocatalysts in action at X-ray free-electron lasers.
Nat. Methods, 14, 2017
|
|
5N4Q
 
 | | Human myelin protein P2, mutant T51P | | Descriptor: | D-MALATE, Myelin P2 protein, PALMITIC ACID, ... | | Authors: | Ruskamo, S, Kursula, P. | | Deposit date: | 2017-02-11 | | Release date: | 2017-08-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Molecular mechanisms of Charcot-Marie-Tooth neuropathy linked to mutations in human myelin protein P2.
Sci Rep, 7, 2017
|
|
5N4M
 
 | | Human myelin protein P2, mutant I43N | | Descriptor: | D-MALATE, Myelin P2 protein, PALMITIC ACID, ... | | Authors: | Ruskamo, S, Kursula, P. | | Deposit date: | 2017-02-11 | | Release date: | 2017-08-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Molecular mechanisms of Charcot-Marie-Tooth neuropathy linked to mutations in human myelin protein P2.
Sci Rep, 7, 2017
|
|
5N4P
 
 | | Human myelin P2, mutant I52T | | Descriptor: | D-MALATE, Myelin P2 protein, PALMITIC ACID, ... | | Authors: | Ruskamo, S, Kursula, P. | | Deposit date: | 2017-02-11 | | Release date: | 2017-08-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.533 Å) | | Cite: | Molecular mechanisms of Charcot-Marie-Tooth neuropathy linked to mutations in human myelin protein P2.
Sci Rep, 7, 2017
|
|
