1MWZ
 
 | | Solution structure of the N-terminal domain of ZntA in the Zn(II)-form | | Descriptor: | ZINC ION, ZntA | | Authors: | Banci, L, Bertini, I, Ciofi-Baffoni, S, Finney, L.A, Outten, C.E, O'Halloran, T.V. | | Deposit date: | 2002-10-01 | | Release date: | 2002-11-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A new zinc-protein coordination site in intracellular metal trafficking: solution structure of the apo and Zn(II) forms of ZntA (46-118)
J.Mol.Biol., 323, 2002
|
|
1P8G
 
 | |
2KMX
 
 | | Solution structure of the nucleotide binding domain of the human Menkes protein in the ATP-bound form | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Copper-transporting ATPase 1 | | Authors: | Banci, L, Bertini, I, Cantini, F, Inagaki, S, Migliardi, M, Rosato, A. | | Deposit date: | 2009-08-05 | | Release date: | 2009-12-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The binding mode of ATP revealed by the solution structure of the N-domain of human ATP7A.
J.Biol.Chem., 2009
|
|
2MMZ
 
 | | Solution structure of the apo form of human glutaredoxin 5 | | Descriptor: | Glutaredoxin-related protein 5, mitochondrial | | Authors: | Banci, L, Brancaccio, D, Ciofi-Baffoni, S, Del Conte, R, Gadepalli, R, Mikolajczyk, M, Neri, S, Piccioli, M, Winkelmann, J. | | Deposit date: | 2014-03-25 | | Release date: | 2014-04-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | [2Fe-2S] cluster transfer in iron-sulfur protein biogenesis.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2JQA
 
 | | Solution structure of apo-DR1885 from Deinococcus radiodurans | | Descriptor: | Hypothetical protein | | Authors: | Banci, L, Bertini, I, Ciofi Baffoni, S, Katsari, E, Katsaros, N, Kubicek, K. | | Deposit date: | 2007-05-30 | | Release date: | 2007-06-12 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | A copper(I) protein possibly involved in the assembly of CuA center of bacterial cytochrome c oxidase
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2JSC
 
 | | NMR structure of the cadmium metal-sensor CMTR from Mycobacterium tuberculosis | | Descriptor: | CADMIUM ION, Transcriptional regulator Rv1994c/MT2050 | | Authors: | Banci, L, Bertini, I, Cantini, F, Ciofi-Baffoni, S, Cavet, J.S, Dennison, C, Graham, A.I, Harvie, D.R, Robinson, N.J, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2007-07-02 | | Release date: | 2007-07-31 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | NMR Structural Analysis of Cadmium Sensing by Winged Helix Repressor CmtR.
J.Biol.Chem., 282, 2007
|
|
2L4D
 
 | | cytochrome c domain of pp3183 protein from Pseudomonas putida | | Descriptor: | HEME C, SCO1/SenC family protein/cytochrome c | | Authors: | Banci, L, Bertini, I, Ciofi-Baffoni, S, Kozyreva, T, Mori, M, Wang, S. | | Deposit date: | 2010-10-04 | | Release date: | 2011-01-26 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Sco proteins are involved in electron transfer processes
J.Biol.Inorg.Chem., 16, 2011
|
|
2KMV
 
 | | Solution structure of the nucleotide binding domain of the human Menkes protein in the ATP-free form | | Descriptor: | Copper-transporting ATPase 1 | | Authors: | Banci, L, Bertini, I, Cantini, F, Inagaki, S, Migliardi, M, Rosato, A. | | Deposit date: | 2009-08-05 | | Release date: | 2009-12-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The binding mode of ATP revealed by the solution structure of the N-domain of human ATP7A.
J.Biol.Chem., 2009
|
|
2KIJ
 
 | | Solution structure of the Actuator domain of the copper-transporting ATPase ATP7A | | Descriptor: | Copper-transporting ATPase 1 | | Authors: | Banci, L, Bertini, I, Cantini, F, Migliardi, M, Nushi, F, Natile, G, Rosato, A. | | Deposit date: | 2009-05-06 | | Release date: | 2009-08-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the actuator domain of ATP7A and ATP7B, the Menkes and Wilson disease proteins
Biochemistry, 48, 2009
|
|
2LD4
 
 | | Solution structure of the N-terminal domain of human anamorsin | | Descriptor: | Anamorsin | | Authors: | Banci, L, Bertini, I, Ciofi-Baffoni, S, Boscaro, F, Chatzi, A, Mikolajczyk, M, Tokatlidis, K, Winkelmann, J. | | Deposit date: | 2011-05-13 | | Release date: | 2011-07-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Anamorsin Is a [2Fe-2S] Cluster-Containing Substrate of the Mia40-Dependent Mitochondrial Protein Trapping Machinery.
Chem.Biol., 18, 2011
|
|
3RE0
 
 | | Crystal structure of human apo Cu,Zn superoxide dismutase (SOD1) complexed with cisplatin | | Descriptor: | Cisplatin, Superoxide dismutase [Cu-Zn] | | Authors: | Bertini, I, Blazevits, O, Calderone, V, Jaifei, M, Vieru, M, Amori, I, Cozzolino, M, Carri, M.T, Banci, L. | | Deposit date: | 2011-04-02 | | Release date: | 2012-04-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Interaction of cisplatin with human superoxide dismutase.
J.Am.Chem.Soc., 134, 2012
|
|
1XTL
 
 | | Crystal structure of P104H mutant of SOD-like protein from Bacillus subtilis. | | Descriptor: | COPPER (II) ION, Hypothetical superoxide dismutase-like protein yojM, ZINC ION | | Authors: | Calderone, V, Mangani, S, Banci, L, Benvenuti, M, Bertini, I, Viezzoli, M.S, Fantoni, A. | | Deposit date: | 2004-10-22 | | Release date: | 2005-10-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | From an Inactive Prokaryotic SOD Homologue to an Active Protein through Site-Directed Mutagenesis.
J.Am.Chem.Soc., 127, 2005
|
|
1XTM
 
 | | Crystal structure of the double mutant Y88H-P104H of a SOD-like protein from Bacillus subtilis. | | Descriptor: | COPPER (II) ION, Hypothetical superoxide dismutase-like protein yojM, ZINC ION | | Authors: | Calderone, V, Mangani, S, Banci, L, Benvenuti, M, Bertini, I, Fantoni, A, Viezzoli, M.S. | | Deposit date: | 2004-10-22 | | Release date: | 2005-10-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | From an Inactive Prokaryotic SOD Homologue to an Active Protein through Site-Directed Mutagenesis.
J.Am.Chem.Soc., 127, 2005
|
|
2LLH
 
 | | NMR structure of Npm1_c70 | | Descriptor: | Nucleophosmin | | Authors: | Banci, L, Bertini, I, Brunori, M, Di Matteo, A, Federici, L, Gallo, A, Lo Sterzo, C, Mori, M. | | Deposit date: | 2011-11-09 | | Release date: | 2012-06-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of Nucleophosmin DNA-binding Domain and Analysis of Its Complex with a G-quadruplex Sequence from the c-MYC Promoter.
J.Biol.Chem., 287, 2012
|
|
2LDI
 
 | |
2LQT
 
 | |
2N6J
 
 | | Solution structure of Zmp1, a zinc-dependent metalloprotease secreted by Clostridium difficile | | Descriptor: | ZINC ION, Zinc metalloprotease Zmp1 | | Authors: | Banci, L, Cantini, F, Scarselli, M, Rubino, J.T, Martinelli, M. | | Deposit date: | 2015-08-24 | | Release date: | 2016-01-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of zinc-bound Zmp1, a zinc-dependent metalloprotease secreted by Clostridium difficile.
J.Biol.Inorg.Chem., 21, 2016
|
|
2LQL
 
 | | Solution structure of CHCH5 | | Descriptor: | Coiled-coil-helix-coiled-coil-helix domain-containing protein 5 | | Authors: | Peruzzini, R, Ciofi-Baffoni, S, Banci, L, Bertini, I. | | Deposit date: | 2012-03-09 | | Release date: | 2012-10-24 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of CHCHD5 and CHCHD7: Two atypical human twin CX(9)C proteins.
J.Struct.Biol., 180, 2012
|
|
8P6U
 
 | | Human carbonic anhydrase II containing 5-fluorotryptophanes | | Descriptor: | BENZOIC ACID, Carbonic anhydrase 2, MERCURIBENZOIC ACID, ... | | Authors: | Pham, L.B.T, Costantino, A, Barbieri, L, Calderone, V, Luchinat, E, Banci, L. | | Deposit date: | 2023-05-30 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Controlling the incorporation of fluorinated amino acids in human cells and its structural impact.
Protein Sci., 33, 2024
|
|
8PHL
 
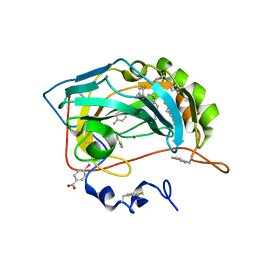 | | Human carbonic anhydrase II containing 4-fluorophenylalanine | | Descriptor: | Carbonic anhydrase 2, MERCURIBENZOIC ACID, ZINC ION | | Authors: | Pham, L.B.T, Costantino, A, Barbieri, L, Calderone, V, Luchinat, E, Banci, L. | | Deposit date: | 2023-06-20 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Controlling the incorporation of fluorinated amino acids in human cells and its structural impact.
Protein Sci., 33, 2024
|
|
8B29
 
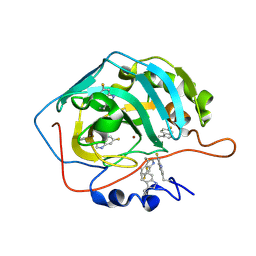 | | Human carbonic anhydrase II containing 6-fluorotryptophanes. | | Descriptor: | Carbonic anhydrase 2, ZINC ION | | Authors: | Pham, L.B.T, Costantino, A, Barbieri, L, Calderone, V, Luchinat, E, Banci, L. | | Deposit date: | 2022-09-13 | | Release date: | 2023-01-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Direct Expression of Fluorinated Proteins in Human Cells for 19 F In-Cell NMR Spectroscopy.
J.Am.Chem.Soc., 145, 2023
|
|
8Q0C
 
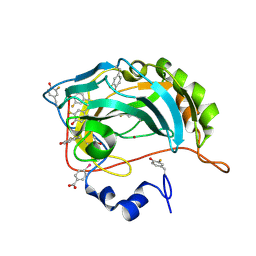 | | Human carbonic anhydrase II containing 3-fluorotyrosine | | Descriptor: | Carbonic anhydrase 2, MERCURIBENZOIC ACID, ZINC ION | | Authors: | Pham, L.B.T, Costantino, A, Barbieri, L, Calderone, V, Luchinat, E, Banci, L. | | Deposit date: | 2023-07-28 | | Release date: | 2024-03-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Controlling the incorporation of fluorinated amino acids in human cells and its structural impact.
Protein Sci., 33, 2024
|
|
6GEU
 
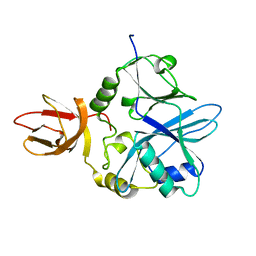 | | Crystal structure of the C230A mutant of human IBA57 | | Descriptor: | Putative transferase CAF17, mitochondrial | | Authors: | Calderone, V, Ciofi-Baffoni, S, Gourdoupis, S, Banci, L, Nasta, V. | | Deposit date: | 2018-04-27 | | Release date: | 2019-04-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | IBA57 Recruits ISCA2 to Form a [2Fe-2S] Cluster-Mediated Complex.
J.Am.Chem.Soc., 140, 2018
|
|
5LCI
 
 | |
8B0T
 
 | | SARS-CoV-2 Main Protease adduct with Au(PEt3)Br | | Descriptor: | 3C-like proteinase nsp5, GOLD ION | | Authors: | Massai, L, Grifagni, D, De Santis, A, Geri, A, Calderone, V, Cantini, F, Banci, L, Messori, L. | | Deposit date: | 2022-09-08 | | Release date: | 2022-12-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Gold-Based Metal Drugs as Inhibitors of Coronavirus Proteins: The Inhibition of SARS-CoV-2 Main Protease by Auranofin and Its Analogs.
Biomolecules, 12, 2022
|
|
