3MD3
 
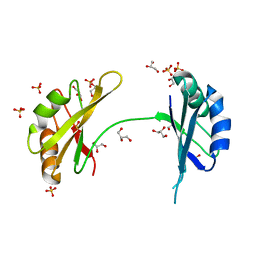 | | Crystal Structure of the First Two RRM Domains of Yeast Poly(U) Binding Protein (Pub1) | | Descriptor: | GLYCEROL, Nuclear and cytoplasmic polyadenylated RNA-binding protein PUB1, SULFATE ION | | Authors: | Li, H, Shi, H, Zhu, Z, Wang, H, Niu, L, Teng, M. | | Deposit date: | 2010-03-29 | | Release date: | 2010-05-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of the First Two RRM Domains of Yeast Poly(U) Binding Protein (Pub1)
To be published
|
|
1NSI
 
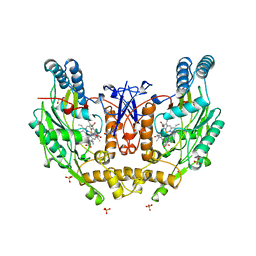 | | HUMAN INDUCIBLE NITRIC OXIDE SYNTHASE, ZN-BOUND, L-ARG COMPLEX | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ARGININE, GLYCEROL, ... | | Authors: | Li, H, Raman, C.S, Glaser, C.B, Blasko, E, Young, T.A, Parkinson, J.F, Whitlow, M, Poulos, T.L. | | Deposit date: | 1999-01-10 | | Release date: | 2000-01-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structures of zinc-free and -bound heme domain of human inducible nitric-oxide synthase. Implications for dimer stability and comparison with endothelial nitric-oxide synthase.
J.Biol.Chem., 274, 1999
|
|
1OM5
 
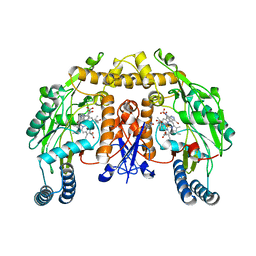 | | STRUCTURE OF RAT NEURONAL NOS HEME DOMAIN WITH 3-BROMO-7-NITROINDAZOLE BOUND | | Descriptor: | 3-BROMO-7-NITROINDAZOLE, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | Authors: | Li, H, Martasek, P, Masters, B.S.S, Poulos, T.L, Raman, C.S. | | Deposit date: | 2003-02-24 | | Release date: | 2003-03-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of Rat Neuronal NOS Heme Domain with 3-Bromo-7-Nitroindazole Bound
To be Published
|
|
1OSP
 
 | |
1V89
 
 | | Solution Structure of the Pleckstrin Homology Domain of Human KIAA0053 Protein | | Descriptor: | Hypothetical protein KIAA0053 | | Authors: | Li, H, Tochio, N, Koshiba, S, Inoue, M, Hirota, H, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-12-29 | | Release date: | 2004-06-29 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Pleckstrin Homology Domain of Human KIAA0053 Protein
To be Published
|
|
1D1W
 
 | | BOVINE ENDOTHELIAL NITRIC OXIDE SYNTHASE HEME DOMAIN COMPLEXED WITH 2-AMINOTHIAZOLINE (H4B BOUND) | | Descriptor: | 2-AMINOTHIAZOLINE, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | Authors: | Li, H, Raman, C.S, Martasek, P, Kral, V, Masters, B.S.S, Poulos, T.L. | | Deposit date: | 1999-09-21 | | Release date: | 2000-10-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mapping the active site polarity in structures of endothelial nitric oxide synthase heme domain complexed with isothioureas.
J.Inorg.Biochem., 81, 2000
|
|
1WWQ
 
 | | Solution Structure of Mouse ER | | Descriptor: | Enhancer of rudimentary homolog | | Authors: | Li, H, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-01-12 | | Release date: | 2006-01-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the mouse enhancer of rudimentary protein reveals a novel fold
J.Biomol.Nmr, 32, 2005
|
|
2YT0
 
 | | Solution structure of the chimera of the C-terminal tail peptide of APP and the C-terminal PID domain of Fe65L | | Descriptor: | Amyloid beta A4 protein and Amyloid beta A4 precursor protein-binding family B member 2 | | Authors: | Li, H, Koshiba, S, Tochio, N, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-05 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the C-terminal phosphotyrosine interaction domain of Fe65L1 complexed with the cytoplasmic tail of amyloid precursor protein reveals a novel peptide binding mode
J.Biol.Chem., 283, 2008
|
|
2YS4
 
 | | Solution structure of the N-terminal PapD-like domain of HYDIN protein from human | | Descriptor: | Hydrocephalus-inducing protein homolog | | Authors: | Li, H, Tomizawa, T, Koshiba, S, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-03 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal PapD-like domain of HYDIN protein from human
To be Published
|
|
2YT1
 
 | | Solution structure of the chimera of the C-terminal tail peptide of APP and the C-terminal PID domain of Fe65L | | Descriptor: | Amyloid beta A4 protein and Amyloid beta A4 precursor protein-binding family B member 2 | | Authors: | Li, H, Koshiba, S, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-05 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the C-terminal phosphotyrosine interaction domain of Fe65L1 complexed with the cytoplasmic tail of amyloid precursor protein reveals a novel peptide binding mode
J.Biol.Chem., 283, 2008
|
|
2BMH
 
 | |
2YS1
 
 | | Solution structure of the PH domain of Dynamin-2 from human | | Descriptor: | Dynamin-2 | | Authors: | Li, H, Sato, M, Tochio, N, Koshiba, S, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-03 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the PH domain of Dynamin-2 from human
To be Published
|
|
2YRY
 
 | | Solution structure of the PH domain of Pleckstrin homology domain-containing family A member 6 from human | | Descriptor: | Pleckstrin homology domain-containing family A member 6 | | Authors: | Li, H, Sato, M, Koshiba, S, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-03 | | Release date: | 2007-10-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the PH domain of Pleckstrin homology domain-containing family A member 6 from human
To be Published
|
|
2YS3
 
 | | Solution structure of the PH domain of Kindlin-3 from human | | Descriptor: | Unc-112-related protein 2 | | Authors: | Li, H, Sato, M, Koshiba, S, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-03 | | Release date: | 2007-10-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the PH domain of Kindlin-3 from human
To be Published
|
|
2YSZ
 
 | | Solution structure of the chimera of the C-terminal PID domain of Fe65L and the C-terminal tail peptide of APP | | Descriptor: | Amyloid beta A4 precursor protein-binding family B member 2 and Amyloid beta A4 protein | | Authors: | Li, H, Koshiba, S, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-05 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the C-terminal phosphotyrosine interaction domain of Fe65L1 complexed with the cytoplasmic tail of amyloid precursor protein reveals a novel peptide binding mode
J.Biol.Chem., 283, 2008
|
|
3E9C
 
 | |
8IRI
 
 | | XFEL structure of cyanobacterial photosystem II following two flashes (2F) with a 5-millisecond delay | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Li, H, Suga, M, Shen, J.R. | | Deposit date: | 2023-03-17 | | Release date: | 2024-01-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Oxygen-evolving photosystem II structures during S 1 -S 2 -S 3 transitions.
Nature, 626, 2024
|
|
3E9D
 
 | | Structure of full-length TIGAR from Danio rerio | | Descriptor: | PHOSPHATE ION, POTASSIUM ION, Zgc:56074 | | Authors: | Li, H, Jogl, G. | | Deposit date: | 2008-08-21 | | Release date: | 2008-12-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | TIGAR (TP53 induced glycolysis and apoptosis regulator) is a fructose-2,6- and fructose-1,6-bisphosphatase
To be Published
|
|
3E9E
 
 | |
8IRG
 
 | | XFEL structure of cyanobacterial photosystem II following two flashes (2F) with a 30-microsecond delay | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Li, H, Suga, M, Shen, J.R. | | Deposit date: | 2023-03-17 | | Release date: | 2024-01-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Oxygen-evolving photosystem II structures during S 1 -S 2 -S 3 transitions.
Nature, 626, 2024
|
|
8IR6
 
 | | XFEL structure of cyanobacterial photosystem II following one flash (1F) with a 20-nanosecond delay | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Li, H, Suga, M, Shen, J.R. | | Deposit date: | 2023-03-17 | | Release date: | 2024-01-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Oxygen-evolving photosystem II structures during S 1 -S 2 -S 3 transitions.
Nature, 626, 2024
|
|
8IR7
 
 | | XFEL structure of cyanobacterial photosystem II following one flash (1F) with a 200-nanosecond delay | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Li, H, Suga, M, Shen, J.R. | | Deposit date: | 2023-03-17 | | Release date: | 2024-01-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Oxygen-evolving photosystem II structures during S 1 -S 2 -S 3 transitions.
Nature, 626, 2024
|
|
8IR8
 
 | | XFEL structure of cyanobacterial photosystem II following one flash (1F) with a 1-microsecond delay | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Li, H, Suga, M, Shen, J.R. | | Deposit date: | 2023-03-17 | | Release date: | 2024-01-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Oxygen-evolving photosystem II structures during S 1 -S 2 -S 3 transitions.
Nature, 626, 2024
|
|
8IRB
 
 | | XFEL structure of cyanobacterial photosystem II following one flash (1F) with a 5-millisecond delay | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Li, H, Suga, M, Shen, J.R. | | Deposit date: | 2023-03-17 | | Release date: | 2024-01-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Oxygen-evolving photosystem II structures during S 1 -S 2 -S 3 transitions.
Nature, 626, 2024
|
|
8IRH
 
 | | XFEL structure of cyanobacterial photosystem II following two flashes (2F) with a 200-microsecond delay | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Li, H, Suga, M, Shen, J.R. | | Deposit date: | 2023-03-17 | | Release date: | 2024-01-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Oxygen-evolving photosystem II structures during S 1 -S 2 -S 3 transitions.
Nature, 626, 2024
|
|
