6UBO
 
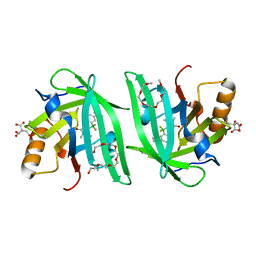 | | Fluorogen Activating Protein Dib1 | | Descriptor: | 12-(diethylamino)-2,2-bis(fluoranyl)-4,5-dimethyl-5-aza-3-azonia-2-boranuidatricyclo[7.4.0.0^{3,7}]trideca-1(13),3,7,9,11-pentaen-6-one, CITRIC ACID, Outer membrane lipoprotein Blc, ... | | Authors: | Muslinkina, L, Pletneva, N, Pletnev, V.Z, Pletnev, S. | | Deposit date: | 2019-09-12 | | Release date: | 2020-09-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structure-Based Rational Design of Two Enhanced Bacterial Lipocalin Blc Tags for Protein-PAINT Super-resolution Microscopy.
Acs Chem.Biol., 15, 2020
|
|
5J76
 
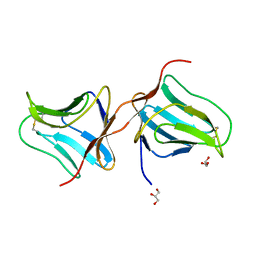 | | Structure of Lectin from Colocasia esculenta(L.) Schott | | Descriptor: | 12kD storage protein, GLYCEROL | | Authors: | Vajravijayan, S, Pletnev, S, Nandhagopal, N, Gunasekaran, K. | | Deposit date: | 2016-04-06 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural analysis of beta-prism lectin from Colocasia esculenta (L.) S chott.
Int.J.Biol.Macromol., 91, 2016
|
|
4ZFS
 
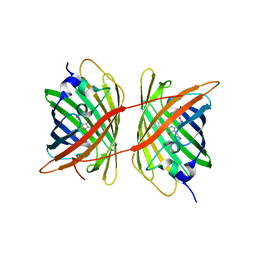 | |
3F2L
 
 | | Crystal structure analysis of the K171A mutation of N-terminal type II cohesin 1 from the cellulosomal ScaB subunit of Acetivibrio cellulolyticus | | Descriptor: | 1,2-ETHANEDIOL, AMMONIUM ION, Cellulosomal scaffoldin adaptor protein B, ... | | Authors: | Frolow, F, Freeman, A, Wine, Y, Eppel, A, Shanzer, M, Stempler, S. | | Deposit date: | 2008-10-30 | | Release date: | 2008-12-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure analysis of the K171A mutation of N-terminal type II cohesin 1 from the cellulosomal ScaB subunit of Acetivibrio cellulolyticus
To be Published
|
|
2FL1
 
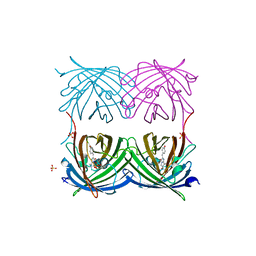 | | Crystal structure of red fluorescent protein from Zoanthus, zRFP574, at 2.4A resolution | | Descriptor: | Red fluorescent protein zoanRFP, SULFATE ION | | Authors: | Pletnev, V, Pletneva, N, Martynov, V, Tikhonova, T, Popov, B, Pletnev, S. | | Deposit date: | 2006-01-05 | | Release date: | 2007-01-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of a red fluorescent protein from Zoanthus, zRFP574, reveals a novel chromophore
Acta Crystallogr.,Sect.D, 62, 2006
|
|
8EEU
 
 | | Venezuelan equine encephalitis virus-like particle in complex with Fab SKT05 | | Descriptor: | Coat protein, Fab SKT05 heavy chain, Fab SKT05 light chain | | Authors: | Tsybovsky, Y, Pletnev, S, Verardi, R, Roederer, M, Kwong, P.D. | | Deposit date: | 2022-09-07 | | Release date: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Vaccine elicitation and structural basis for antibody protection against alphaviruses.
Cell, 186, 2023
|
|
8EEV
 
 | | Venezuelan equine encephalitis virus-like particle in complex with Fab SKT-20 | | Descriptor: | Coat protein, Fab SKT20 heavy chain, Fab SKT20 light chain | | Authors: | Tsybovsky, Y, Pletnev, S, Verardi, R, Roederer, M, Kwong, P.D. | | Deposit date: | 2022-09-07 | | Release date: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Vaccine elicitation and structural basis for antibody protection against alphaviruses.
Cell, 186, 2023
|
|
6LIK
 
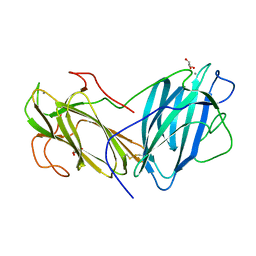 | | Crystal Structure of Lectin from Pleurotus ostreatus in complex with Galactose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Vajravijayan, S, Pletnev, S, Luo, Z, Iqbal, S, Nandhagopal, N, Gunasekaran, K. | | Deposit date: | 2019-12-11 | | Release date: | 2020-08-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystallographic and calorimetric analysis on Pleurotus ostreatus lectin and its sugar complexes - promiscuous binding driven by geometry.
Int.J.Biol.Macromol., 152, 2020
|
|
4ORN
 
 | | Blue Fluorescent Protein mKalama1 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, SULFATE ION, ... | | Authors: | Vegh, R.B, Bloch, D.A, Bommarius, A.S, Verkhovsky, M, Pletnev, S, Iwai, H, Bochenkova, A.V, Solntsev, K.M. | | Deposit date: | 2014-02-11 | | Release date: | 2015-02-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Hidden photoinduced reactivity of the blue fluorescent protein mKalama1.
Phys Chem Chem Phys, 17, 2015
|
|
6KC2
 
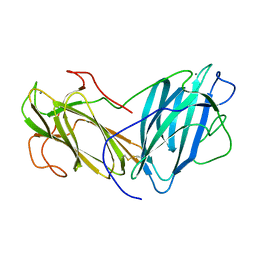 | | Crystal Structure of Lectin from Pleurotus ostreatus in complex with Rhamnose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Lectin, ... | | Authors: | Gunasekaran, K, Pletnev, S, Luo, Z, Vajravijayan, S, Nandhagopal, N. | | Deposit date: | 2019-06-26 | | Release date: | 2020-08-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.246 Å) | | Cite: | Crystallographic and calorimetric analysis on Pleurotus ostreatus lectin and its sugar complexes - promiscuous binding driven by geometry.
Int.J.Biol.Macromol., 152, 2020
|
|
6KBQ
 
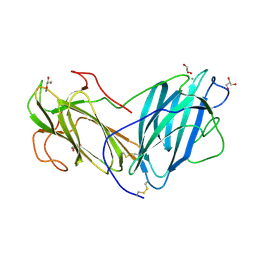 | | Crystal Structure of Lectin from Pleurotus ostreatus in complex with Glycerol | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Vajravijayan, S, Pletnev, S, Luo, Z, Gunasekaran, K, Nandhagopal, N. | | Deposit date: | 2019-06-26 | | Release date: | 2020-08-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Crystallographic and calorimetric analysis on Pleurotus ostreatus lectin and its sugar complexes - promiscuous binding driven by geometry.
Int.J.Biol.Macromol., 152, 2020
|
|
6KBJ
 
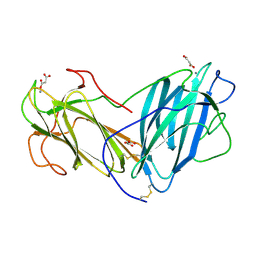 | | Structure of Lectin from Pleurotus ostreatus in complex with malonate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Lectin, ... | | Authors: | Vajravijayan, S, Pletnev, S, Luo, Z, Gunasekaran, K, Nandhagopal, N. | | Deposit date: | 2019-06-25 | | Release date: | 2020-08-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallographic and calorimetric analysis on Pleurotus ostreatus lectin and its sugar complexes - promiscuous binding driven by geometry.
Int.J.Biol.Macromol., 152, 2020
|
|
6LI7
 
 | | Crystal Structure of Lectin from Pleurotus ostreatus in complex with GalNAc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Vajravijayan, S, Pletnev, S, Luo, Z, Ankur, T, Gunasekaran, K, Nandhagopal, N. | | Deposit date: | 2019-12-10 | | Release date: | 2020-08-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.098 Å) | | Cite: | Crystallographic and calorimetric analysis on Pleurotus ostreatus lectin and its sugar complexes - promiscuous binding driven by geometry.
Int.J.Biol.Macromol., 152, 2020
|
|
6M9X
 
 | |
6MAS
 
 | | X-ray Structure of Branchiostoma floridae fluorescent protein lanFP10G | | Descriptor: | GLYCEROL, Uncharacterized protein | | Authors: | Muslinkina, L, Pletneva, N, Pletnev, V, Pletnev, S. | | Deposit date: | 2018-08-28 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural Factors Enabling Successful GFP-Like Proteins with Alanine as the Third Chromophore-Forming Residue.
J. Mol. Biol., 431, 2019
|
|
6M9Z
 
 | |
6M9Y
 
 | |
5WQU
 
 | | Crystal structure of Sweet Potato Beta-Amylase complexed with Maltotetraose | | Descriptor: | Beta-amylase, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Vajravijayan, S, Sergei, P, Nandhagopal, N, Gunasekaran, K. | | Deposit date: | 2016-11-28 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural insights on starch hydrolysis by plant beta-amylase and its evolutionary relationship with bacterial enzymes
Int. J. Biol. Macromol., 113, 2018
|
|
5WQS
 
 | | Crystal structure of Apo Beta-Amylase from Sweet potato | | Descriptor: | Beta-amylase, ISOPROPYL ALCOHOL | | Authors: | Vajravijayan, S, Sergei, P, Nandhagopal, N, Gunasekaran, K. | | Deposit date: | 2016-11-28 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights on starch hydrolysis by plant beta-amylase and its evolutionary relationship with bacterial enzymes
Int. J. Biol. Macromol., 113, 2018
|
|
3LVD
 
 | |
3LVC
 
 | |
3LVA
 
 | |
2GFS
 
 | | P38 Kinase Crystal Structure in complex with RO3201195 | | Descriptor: | Mitogen-Activated Protein Kinase 14, [5-AMINO-1-(4-FLUOROPHENYL)-1H-PYRAZOL-4-YL](3-{[(2R)-2,3-DIHYDROXYPROPYL]OXY}PHENYL)METHANONE | | Authors: | Harris, S.F, Bertrand, J, Villasenor, A. | | Deposit date: | 2006-03-23 | | Release date: | 2006-04-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Discovery of S-[5-Amino-1-(4-fluorophenyl)-1H-pyrazol-4-yl]-[3-(2,3-dihydroxypropoxy)phenyl]-methanone (RO3201195), and Orally Bioavailable and Highly Selective Inhibitor of p38 Map Kinase
J.Med.Chem., 49, 2006
|
|
6ZQO
 
 | | EYFP mutant - F165G | | Descriptor: | G protein/GFP fusion protein, SULFATE ION | | Authors: | Pletnev, V.Z, Pletnev, S.V, Pletneva, N.V. | | Deposit date: | 2020-07-10 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Amino acid residue at the 165th position tunes EYFP chromophore maturation. A structure-based design.
Comput Struct Biotechnol J, 19, 2021
|
|
4ZBL
 
 | |
