7ZDG
 
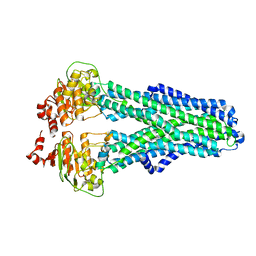 | | IF(heme/confined) conformation of CydDC (Dataset-5) | | Descriptor: | ATP-binding/permease protein CydC, ATP-binding/permease protein CydD, HEME B/C | | Authors: | Wu, D, Safarian, S. | | Deposit date: | 2022-03-29 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Dissecting the conformational complexity and mechanism of a bacterial heme transporter.
Nat.Chem.Biol., 19, 2023
|
|
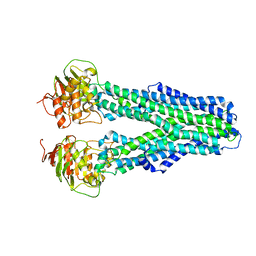
7ZDS
 
 | |
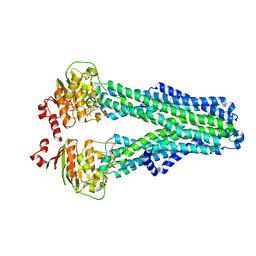
7ZDV
 
 | |
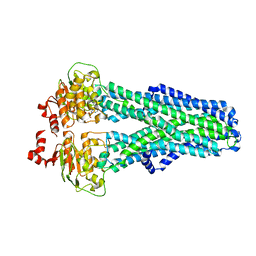
7ZDF
 
 | |
7ZDT
 
 | |
7ZDE
 
 | |
7ZEC
 
 | |
7ZDW
 
 | |
7ZDL
 
 | |
7ZE5
 
 | |
7ZDC
 
 | |
7ZDK
 
 | |
7ZD5
 
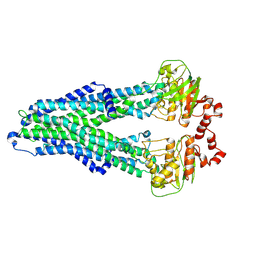 | | IF(apo/as isolated) conformation of CydDC (Dataset-1) | | Descriptor: | ATP-binding/permease protein CydC, ATP-binding/permease protein CydD | | Authors: | Wu, D, Safarian, S. | | Deposit date: | 2022-03-29 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Dissecting the conformational complexity and mechanism of a bacterial heme transporter.
Nat.Chem.Biol., 19, 2023
|
|
7ZDU
 
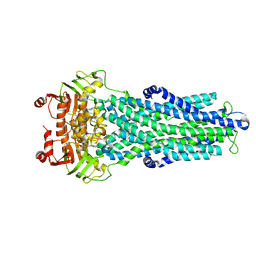 | |
7YMV
 
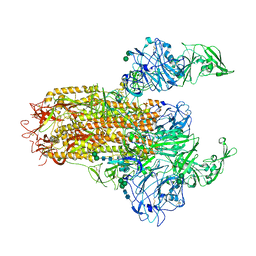 | | Cryo-EM structure of MERS-CoV spike protein, Two RBD-up conformation 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hsu, S.T.D, Chang, N.E, Weng, Z.W, Yang, T.J, Draczkowski, P. | | Deposit date: | 2022-07-29 | | Release date: | 2023-08-09 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (6.74 Å) | | Cite: | Rapid simulation of glycoprotein structures by grafting and steric exclusion of glycan conformer libraries.
Cell, 187, 2024
|
|
7YMW
 
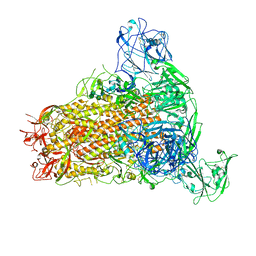 | | Cryo-EM structure of MERS-CoV spike protein, One RBD-up conformation 4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Hsu, S.T.D, Chang, N.E, Weng, Z.W, Yang, T.J, Draczkowski, P. | | Deposit date: | 2022-07-29 | | Release date: | 2023-08-09 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (6.05 Å) | | Cite: | Rapid simulation of glycoprotein structures by grafting and steric exclusion of glycan conformer libraries.
Cell, 187, 2024
|
|
7YMX
 
 | | Cryo-EM structure of MERS-CoV spike protein, One RBD-up conformation 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Hsu, S.T.D, Chang, N.E, Weng, Z.W, Yang, T.J, Draczkowski, P. | | Deposit date: | 2022-07-29 | | Release date: | 2023-08-09 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (4.44 Å) | | Cite: | Rapid simulation of glycoprotein structures by grafting and steric exclusion of glycan conformer libraries.
Cell, 187, 2024
|
|
7YMT
 
 | | Cryo-EM structure of MERS-CoV spike protein, Two RBD-up conformation 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Hsu, S.T.D, Chang, N.E, Weng, Z.W, Yang, T.J, Draczkowski, P. | | Deposit date: | 2022-07-29 | | Release date: | 2023-08-09 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (6.55 Å) | | Cite: | Rapid simulation of glycoprotein structures by grafting and steric exclusion of glycan conformer libraries.
Cell, 187, 2024
|
|
7YN0
 
 | | Cryo-EM structure of MERS-CoV spike protein, all RBD-down conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hsu, S.T.D, Chang, N.E, Weng, Z.W, Yang, T.J, Draczkowski, P. | | Deposit date: | 2022-07-29 | | Release date: | 2023-08-09 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Rapid simulation of glycoprotein structures by grafting and steric exclusion of glycan conformer libraries.
Cell, 187, 2024
|
|
7YMY
 
 | | Cryo-EM structure of MERS-CoV spike protein, One RBD-up conformation 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Hsu, S.T.D, Chang, N.E, Weng, Z.W, Yang, T.J, Draczkowski, P. | | Deposit date: | 2022-07-29 | | Release date: | 2023-08-09 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (4.96 Å) | | Cite: | Rapid simulation of glycoprotein structures by grafting and steric exclusion of glycan conformer libraries.
Cell, 187, 2024
|
|
7YMZ
 
 | | Cryo-EM structure of MERS-CoV spike protein, intermediate conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Hsu, S.T.D, Chang, N.E, Weng, Z.W, Yang, T.J, Draczkowski, P. | | Deposit date: | 2022-07-29 | | Release date: | 2023-08-09 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (4.39 Å) | | Cite: | Rapid simulation of glycoprotein structures by grafting and steric exclusion of glycan conformer libraries.
Cell, 187, 2024
|
|
6ZH0
 
 | | Structure of human galactokinase 1 bound with 2-(4-chlorophenyl)-N-(pyrimidin-2-yl)acetamide | | Descriptor: | 2-(1,3-benzoxazol-2-ylamino)spiro[1,6,7,8-tetrahydroquinazoline-4,1'-cyclohexane]-5-one, Galactokinase, N-(3-chlorophenyl)-2,2,2-trifluoroacetamide, ... | | Authors: | Mackinnon, S.R, Bezerra, G.A, Zhang, M, Foster, W, Krojer, T, Brandao-Neto, J, Douangamath, A, Arrowsmith, C, Edwards, A, Bountra, C, Brennan, P, Lai, K, Yue, W.W. | | Deposit date: | 2020-06-20 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Fragment Screening Reveals Starting Points for Rational Design of Galactokinase 1 Inhibitors to Treat Classic Galactosemia.
Acs Chem.Biol., 16, 2021
|
|
2PJW
 
 | |
5D5R
 
 | | Horse-heart myoglobin - deoxy state | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Barends, T, Schlichting, I. | | Deposit date: | 2015-08-11 | | Release date: | 2015-09-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Direct observation of ultrafast collective motions in CO myoglobin upon ligand dissociation.
Science, 350, 2015
|
|
8QCT
 
 | |
