7U2T
 
 | |
7U2Q
 
 | |
6GG2
 
 | | The structure of FsqB from Aspergillus fumigatus, a flavoenzyme of the amine oxidase family | | Descriptor: | Amino acid oxidase fmpA, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Pavkov-Keller, T, Lahham, M, Macheroux, P, Gruber, K. | | Deposit date: | 2018-05-02 | | Release date: | 2018-09-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.598 Å) | | Cite: | Oxidative cyclization ofN-methyl-dopa by a fungal flavoenzyme of the amine oxidase family.
J. Biol. Chem., 293, 2018
|
|
8AYQ
 
 | | NaK C-DI mutant with Rb+ and Ca2+ | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Minniberger, S, Plested, A.J.R. | | Deposit date: | 2022-09-02 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Mechanism of Calcium Permeation in a Glutamate Receptor Ion Channel.
J.Chem.Inf.Model., 63, 2023
|
|
8AYP
 
 | | NaK C-DI mutant with Rb+ and Ba2+ | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, BARIUM ION, Potassium channel protein, ... | | Authors: | Minniberger, S, Plested, A.J.R. | | Deposit date: | 2022-09-02 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanism of Calcium Permeation in a Glutamate Receptor Ion Channel.
J.Chem.Inf.Model., 63, 2023
|
|
9AV0
 
 | | Crystal structure of S. aureus GuaB dCBS with inhibitor GNE2011 | | Descriptor: | 9-{(1R)-1-[(5P)-5-(4-chloro-1H-imidazol-2-yl)pyridin-3-yl]ethoxy}-1,4-dihydro-2H-pyrano[3,4-c]quinoline, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase | | Authors: | Harris, S.F, Wu, P. | | Deposit date: | 2024-03-01 | | Release date: | 2024-11-27 | | Last modified: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of GuaB inhibitors with efficacy against Acinetobacter baumannii infection.
Mbio, 15, 2024
|
|
9AUX
 
 | | Crystal structure of A. baumannii GuaB dCBS with inhibitor GNE2011 | | Descriptor: | 9-{(1R)-1-[(5P)-5-(4-chloro-1H-imidazol-2-yl)pyridin-3-yl]ethoxy}-1,4-dihydro-2H-pyrano[3,4-c]quinoline, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase | | Authors: | Harris, S.F, Wu, P. | | Deposit date: | 2024-03-01 | | Release date: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Discovery of GuaB inhibitors with efficacy against Acinetobacter baumannii infection.
Mbio, 15, 2024
|
|
9AV2
 
 | | Crystal structure of E. coli GuaB dCBS with inhibitor GNE9979 | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase, N-[4-chloro-3-(dimethylamino)phenyl]-N~2~-[3-(hydroxymethyl)quinolin-6-yl]-L-alaninamide | | Authors: | Harris, S.F, Wu, P. | | Deposit date: | 2024-03-01 | | Release date: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of GuaB inhibitors with efficacy against Acinetobacter baumannii infection.
Mbio, 15, 2024
|
|
9AV1
 
 | | Crystal structure of E. coli GuaB dCBS with inhibitor GNE9123 | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase, N-(6-chloropyridin-3-yl)-N~2~-(1,4-dihydro-2H-pyrano[3,4-c]quinolin-9-yl)-L-alaninamide | | Authors: | Harris, S.F, Wu, P. | | Deposit date: | 2024-03-01 | | Release date: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of GuaB inhibitors with efficacy against Acinetobacter baumannii infection.
Mbio, 15, 2024
|
|
9AUW
 
 | | Crystal structure of A. baumannii GuaB dCBS with inhibitor GNE9979 | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase, N-[4-chloro-3-(dimethylamino)phenyl]-N~2~-[3-(hydroxymethyl)quinolin-6-yl]-L-alaninamide | | Authors: | Harris, S.F, Wu, P. | | Deposit date: | 2024-03-01 | | Release date: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of GuaB inhibitors with efficacy against Acinetobacter baumannii infection.
Mbio, 15, 2024
|
|
9AV3
 
 | | Crystal structure of E. coli GuaB dCBS with inhibitor GNE2011 | | Descriptor: | 9-{(1R)-1-[(5P)-5-(4-chloro-1H-imidazol-2-yl)pyridin-3-yl]ethoxy}-1,4-dihydro-2H-pyrano[3,4-c]quinoline, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase | | Authors: | Harris, S.F, Wu, P. | | Deposit date: | 2024-03-01 | | Release date: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Discovery of GuaB inhibitors with efficacy against Acinetobacter baumannii infection.
Mbio, 15, 2024
|
|
9AUV
 
 | | Crystal structure of A. baumannii GuaB dCBS with inhibitor GNE9123 | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase, N-(6-chloropyridin-3-yl)-N~2~-(1,4-dihydro-2H-pyrano[3,4-c]quinolin-9-yl)-L-alaninamide | | Authors: | Harris, S.F, Wu, P. | | Deposit date: | 2024-03-01 | | Release date: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Discovery of GuaB inhibitors with efficacy against Acinetobacter baumannii infection.
Mbio, 15, 2024
|
|
9AUY
 
 | | Crystal structure of S. aureus GuaB dCBS with inhibitor GNE9123 | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase, N-(6-chloropyridin-3-yl)-N~2~-(1,4-dihydro-2H-pyrano[3,4-c]quinolin-9-yl)-L-alaninamide | | Authors: | Harris, S.F, Wu, P. | | Deposit date: | 2024-03-01 | | Release date: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Discovery of GuaB inhibitors with efficacy against Acinetobacter baumannii infection.
Mbio, 15, 2024
|
|
9AUZ
 
 | | Crystal structure of S. aureus GuaB dCBS with inhibitor GNE9979 | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase, N-[4-chloro-3-(dimethylamino)phenyl]-N~2~-[3-(hydroxymethyl)quinolin-6-yl]-L-alaninamide | | Authors: | Harris, S.F, Wu, P. | | Deposit date: | 2024-03-01 | | Release date: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Discovery of GuaB inhibitors with efficacy against Acinetobacter baumannii infection.
Mbio, 15, 2024
|
|
8Q3I
 
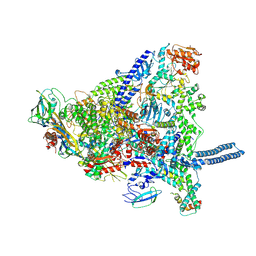 | | Mycobacterium smegmatis RNA polymerase in complex with HelD, SigA and RbpA in State I | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Koval, T, Krasny, L, Dohnalek, J, Kouba, T. | | Deposit date: | 2023-08-04 | | Release date: | 2024-08-14 | | Last modified: | 2024-12-11 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Mycobacterial HelD connects RNA polymerase recycling with transcription initiation.
Nat Commun, 15, 2024
|
|
8QN8
 
 | | Mycobacterium smegmatis RNA polymerase in complex with HelD, SigA and RbpA in State II | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Koval, T, Krasny, L, Dohnalek, J, Kouba, T. | | Deposit date: | 2023-09-25 | | Release date: | 2024-10-02 | | Last modified: | 2024-12-11 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Mycobacterial HelD connects RNA polymerase recycling with transcription initiation.
Nat Commun, 15, 2024
|
|
8QTI
 
 | | Mycobacterium smegnatis RNAP open promoter complex with SigmaA and RbpA | | Descriptor: | DNA 50-mer non-template strand, DNA 50-mer template strand, DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Koval, T, Krasny, L, Dohnalek, J, Kouba, T. | | Deposit date: | 2023-10-12 | | Release date: | 2024-10-02 | | Last modified: | 2024-12-11 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Mycobacterial HelD connects RNA polymerase recycling with transcription initiation.
Nat Commun, 15, 2024
|
|
8QU6
 
 | | Mycobacterium smegnatis RNA polymerase transcription initiation complex with SigmaA, RbpA, HelD and an upstream-fork promoter fragment | | Descriptor: | DNA 22-MER, DNA 31-MER, DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Koval, T, Krasny, L, Dohnalek, J, Kouba, T. | | Deposit date: | 2023-10-13 | | Release date: | 2024-10-02 | | Last modified: | 2024-12-11 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Mycobacterial HelD connects RNA polymerase recycling with transcription initiation.
Nat Commun, 15, 2024
|
|
8R6R
 
 | | Mycobacterium smegnatis RNA polymerase RP2-like transcription initiation complex with SigmaA, RbpA and open promoter DNA | | Descriptor: | DNA 50-MER, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Koval, T, Krasny, L, Dohnalek, J, Kouba, T. | | Deposit date: | 2023-11-22 | | Release date: | 2024-10-02 | | Last modified: | 2024-12-11 | | Method: | ELECTRON MICROSCOPY (3.89 Å) | | Cite: | Mycobacterial HelD connects RNA polymerase recycling with transcription initiation.
Nat Commun, 15, 2024
|
|
8R6P
 
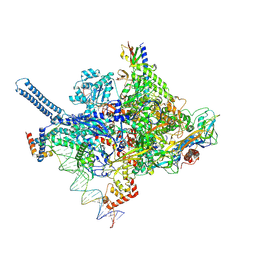 | | Mycobacterium smegnatis RNA polymerase RP2-like transcription initiation complex with SigmaA, RbpA, HelD N-terminal domain and open promoter DNA | | Descriptor: | DNA 50-MER, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Koval, T, Krasny, L, Dohnalek, J, Kouba, T. | | Deposit date: | 2023-11-22 | | Release date: | 2024-10-02 | | Last modified: | 2024-12-11 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Mycobacterial HelD connects RNA polymerase recycling with transcription initiation.
Nat Commun, 15, 2024
|
|
8R2M
 
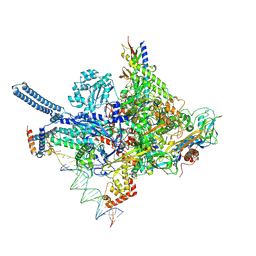 | | Mycobacterium smegnatis RNA polymerase transcription initiation complex with SigmaA, RbpA, HelD N-terminal domain and an upstream-fork promoter fragment; State III conformation | | Descriptor: | DNA 22-MER, DNA 31-MER, DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Koval, T, Krasny, L, Dohnalek, J, Kouba, T. | | Deposit date: | 2023-11-06 | | Release date: | 2024-10-02 | | Last modified: | 2024-12-11 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Mycobacterial HelD connects RNA polymerase recycling with transcription initiation.
Nat Commun, 15, 2024
|
|
8R3M
 
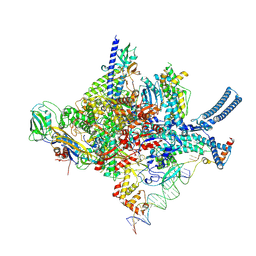 | | Mycobacterium smegnatis RNA polymerase transcription initiation complex with SigmaA, RbpA, HelD N-terminal, CO and PCh loop domain, and an upstream-fork promoter fragment; State III conformation | | Descriptor: | DNA 22-MER, DNA 31-MER, DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Koval, T, Krasny, L, Dohnalek, J, Kouba, T. | | Deposit date: | 2023-11-09 | | Release date: | 2024-10-02 | | Last modified: | 2024-12-11 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Mycobacterial HelD connects RNA polymerase recycling with transcription initiation.
Nat Commun, 15, 2024
|
|
6XLP
 
 | | Structure of the essential inner membrane lipopolysaccharide-PbgA complex | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-deoxy-3-O-[(1R,3R)-1,3-dihydroxytetradecyl]-2-{[(3R)-3-hydroxytetradecanoyl]amino}-1-O-phosphono-alpha-D-glucopyranose-(6-1)-[3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-6)]1,5-anhydro-2-deoxy-2-{[(1S,3R)-1-hydroxy-3-(pentanoyloxy)undecyl]amino}-4-O-phosphono-D-glucitol, ... | | Authors: | Payandeh, J, Clairefeuille, T. | | Deposit date: | 2020-06-29 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the essential inner membrane lipopolysaccharide-PbgA complex.
Nature, 584, 2020
|
|
9BK6
 
 | | Structure of B10_CYTX binder-complex | | Descriptor: | B10_CYTX binder, Cytotoxin 1 | | Authors: | Bera, A.K, Torres, S.V, Kang, A, Baker, D. | | Deposit date: | 2024-04-26 | | Release date: | 2024-10-09 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | De novo designed proteins neutralize lethal snake venom toxins.
Nature, 639, 2025
|
|
9BK7
 
 | |
