1O8C
 
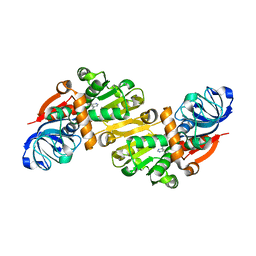 | | CRYSTAL STRUCTURE OF E. COLI K-12 YHDH WITH BOUND NADPH | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, YHDH | | Authors: | Sulzenbacher, G, Roig-Zamboni, V, Pagot, F, Grisel, S, Salamoni, A, Valencia, C, Bignon, C, Vincentelli, R, Tegoni, M, Cambillau, C. | | Deposit date: | 2002-11-26 | | Release date: | 2004-05-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Escherichia Coli Yhdh, a Putative Quinone Oxidoreductase
Acta Crystallogr.,Sect.D, 60, 2004
|
|
7PZZ
 
 | | Crystal structure of serine hydroxymethyltransferase, isoform 2 from Arabidopsis thaliana (SHM2) | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Ruszkowski, M, Sekula, B. | | Deposit date: | 2021-10-13 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Arabidopsis thaliana serine hydroxymethyltransferases: functions, structures, and perspectives.
Plant Physiol Biochem., 187, 2022
|
|
7Q00
 
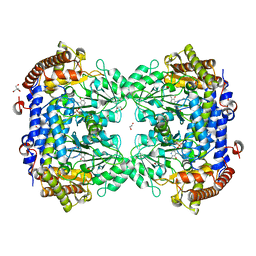 | | Crystal structure of serine hydroxymethyltransferase, isoform 4 from Arabidopsis thaliana (SHM4) | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Serine hydroxymethyltransferase 4 | | Authors: | Ruszkowski, M, Sekula, B. | | Deposit date: | 2021-10-13 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Arabidopsis thaliana serine hydroxymethyltransferases: functions, structures, and perspectives.
Plant Physiol Biochem., 187, 2022
|
|
7QPE
 
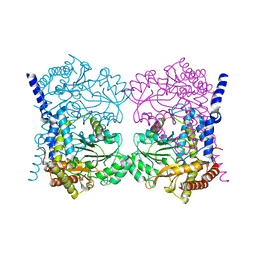 | | Crystal structure of serine hydroxymethyltransferase, isoform 6 from Arabidopsis thaliana (SHM6) | | Descriptor: | NITRATE ION, Serine hydroxymethyltransferase 6 | | Authors: | Ruszkowski, M, Grzechowiak, M, Sekula, B. | | Deposit date: | 2022-01-04 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Arabidopsis thaliana serine hydroxymethyltransferases: functions, structures, and perspectives.
Plant Physiol Biochem., 187, 2022
|
|
7QX8
 
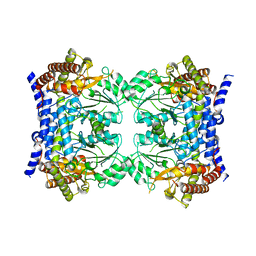 | | Crystal structure of serine hydroxymethyltransferase, isoform 7 from Arabidopsis thaliana (SHM7) | | Descriptor: | Serine hydroxymethyltransferase 7 | | Authors: | Ruszkowski, M, Grzechowiak, M, Sekula, B. | | Deposit date: | 2022-01-26 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Arabidopsis thaliana serine hydroxymethyltransferases: functions, structures, and perspectives.
Plant Physiol Biochem., 187, 2022
|
|
8T1R
 
 | | Crystal structure of human CPSF73 catalytic segment in complex with compound 2 | | Descriptor: | 3-[7,7-bis(oxidanyl)-8-oxa-7-boranuidabicyclo[4.3.0]nona-1,3,5-trien-5-yl]-~{N}-[3-(3-methoxyphenyl)phenyl]propanamide, CHLORIDE ION, Cleavage and polyadenylation specificity factor subunit 3, ... | | Authors: | Huang, J, Tong, L. | | Deposit date: | 2023-06-02 | | Release date: | 2023-11-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Anticancer benzoxaboroles block pre-mRNA processing by directly inhibiting CPSF3.
Cell Chem Biol, 31, 2024
|
|
8T1Q
 
 | | Crystal structure of human CPSF73 catalytic segment in complex with compound 1 | | Descriptor: | 3-[7,7-bis(oxidanyl)-8-oxa-7-boranuidabicyclo[4.3.0]nona-1,3,5-trien-5-yl]-~{N}-[3-(4-ethanoylphenyl)phenyl]propanamide, CHLORIDE ION, Cleavage and polyadenylation specificity factor subunit 3, ... | | Authors: | Huang, J, Tong, L. | | Deposit date: | 2023-06-02 | | Release date: | 2023-11-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Anticancer benzoxaboroles block pre-mRNA processing by directly inhibiting CPSF3.
Cell Chem Biol, 31, 2024
|
|
5AGC
 
 | | Crystallographic forms of the Vps75 tetramer | | Descriptor: | VACUOLAR PROTEIN SORTING-ASSOCIATED PROTEIN 75 | | Authors: | Hammond, C.M, Sundaramoorthy, R, Owen-Hughes, T. | | Deposit date: | 2015-01-29 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | The Histone Chaperone Vps75 Forms Multiple Oligomeric Assemblies Capable of Mediating Exchange between Histone H3-H4 Tetramers and Asf1-H3-H4 Complexes.
Nucleic Acids Res., 44, 2016
|
|
1OJ7
 
 | | STRUCTURAL GENOMICS, UNKNOWN FUNCTION CRYSTAL STRUCTURE OF E. COLI K-12 YQHD | | Descriptor: | 5,6-DIHYDROXY-NADP, BORIC ACID, CHLORIDE ION, ... | | Authors: | Sulzenbacher, G, Perrier, S, Roig-Zamboni, V, Pagot, F, Grisel, S, Salamoni, A, Valencia, C, Bignon, C, Vincentelli, R, Tegoni, M, Cambillau, C. | | Deposit date: | 2003-07-03 | | Release date: | 2004-07-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of E.Coli Alcohol Dehydrogenase Yqhd: Evidence of a Covalently Modified Nadp Coenzyme
J.Mol.Biol., 342, 2004
|
|
4NUT
 
 | | Crystal structure of the complex between Snu13p and the PEP domain of Rsa1 | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, Ribosome assembly 1 protein, SULFATE ION | | Authors: | Charron, C, Chagot, M.E, Manival, X, Branlant, C, Charpentier, B. | | Deposit date: | 2013-12-04 | | Release date: | 2014-12-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Proteomic and 3D structure analyses highlight the C/D box snoRNP assembly mechanism and its control
J.Cell Biol., 207, 2014
|
|
