6SJ2
 
 | | Amidohydrolase, AHS with 3-HAA | | Descriptor: | 3-HYDROXYANTHRANILIC ACID, Amidohydrolase, GLYCEROL, ... | | Authors: | Naismith, J.H, Song, H. | | Deposit date: | 2019-08-12 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | The Biosynthesis of the Benzoxazole in Nataxazole Proceeds via an Unstable Ester and has Synthetic Utility.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6SJ1
 
 | | Amidohydrolase, AHS | | Descriptor: | Amidohydrolase, ZINC ION | | Authors: | Naismith, J.H, Song, H. | | Deposit date: | 2019-08-12 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | The Biosynthesis of the Benzoxazole in Nataxazole Proceeds via an Unstable Ester and has Synthetic Utility.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6SSG
 
 | | Transaminase with DCS bound | | Descriptor: | ForI-DCS, SULFATE ION, [4-[(~{Z})-[(2~{R},5~{R})-5-(azanyloxymethyl)-3,6-bis(oxidanylidene)piperazin-2-yl]methoxyiminomethyl]-6-methyl-5-oxidanyl-pyridin-3-yl]methyl dihydrogen phosphate | | Authors: | Naismith, J.H, Gao, S. | | Deposit date: | 2019-09-06 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | PMP-diketopiperazine adducts form at the active site of a PLP dependent enzyme involved in formycin biosynthesis.
Chem.Commun.(Camb.), 55, 2019
|
|
6SIW
 
 | | PaaK family AMP-ligase with AMP | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Naismith, J.H, Song, H. | | Deposit date: | 2019-08-12 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | The Biosynthesis of the Benzoxazole in Nataxazole Proceeds via an Unstable Ester and has Synthetic Utility.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6SIY
 
 | | PaaK family AMP-ligase with AMP and substrate | | Descriptor: | 1,2-ETHANEDIOL, 3-HYDROXYANTHRANILIC ACID, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Naismith, J.H, Song, H. | | Deposit date: | 2019-08-12 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Biosynthesis of the Benzoxazole in Nataxazole Proceeds via an Unstable Ester and has Synthetic Utility.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6SJ0
 
 | | Amidohydrolase, AHS | | Descriptor: | Amidohydrolase, BICARBONATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Naismith, J.H, Song, H. | | Deposit date: | 2019-08-12 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Biosynthesis of the Benzoxazole in Nataxazole Proceeds via an Unstable Ester and has Synthetic Utility.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6SIZ
 
 | | PaaK family AMP-ligase with ANP and substrate | | Descriptor: | 1,2-ETHANEDIOL, 3-HYDROXYANTHRANILIC ACID, AMP-dependent synthetase and ligase, ... | | Authors: | Naismith, J.H, Song, H. | | Deposit date: | 2019-08-12 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | The Biosynthesis of the Benzoxazole in Nataxazole Proceeds via an Unstable Ester and has Synthetic Utility.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6SJ3
 
 | | Amidohydrolase, AHS with 3-HBA | | Descriptor: | 1,2-ETHANEDIOL, 3-HYDROXYBENZOIC ACID, Amidohydrolase, ... | | Authors: | Naismith, J.H, Song, H. | | Deposit date: | 2019-08-12 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | The Biosynthesis of the Benzoxazole in Nataxazole Proceeds via an Unstable Ester and has Synthetic Utility.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6SSE
 
 | | Transaminase with PMP bound | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, ForI-PMP, SULFATE ION | | Authors: | Naismith, J.H, Gao, S. | | Deposit date: | 2019-09-06 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | PMP-diketopiperazine adducts form at the active site of a PLP dependent enzyme involved in formycin biosynthesis.
Chem.Commun.(Camb.), 55, 2019
|
|
6SIX
 
 | | PaaK family AMP-ligase with ANP | | Descriptor: | 1,2-ETHANEDIOL, AMP-dependent synthetase and ligase, CITRATE ANION, ... | | Authors: | Naismith, J.H, Song, H. | | Deposit date: | 2019-08-12 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | The Biosynthesis of the Benzoxazole in Nataxazole Proceeds via an Unstable Ester and has Synthetic Utility.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6SSF
 
 | | Transaminase with LCS bound | | Descriptor: | ForI-LCS, SULFATE ION, [4-[(~{Z})-[(2~{S},5~{S})-5-(azanyloxymethyl)-3,6-bis(oxidanylidene)piperazin-2-yl]methoxyiminomethyl]-6-methyl-5-oxidanyl-pyridin-3-yl]methyl dihydrogen phosphate | | Authors: | Naismith, J.H, Gao, S. | | Deposit date: | 2019-09-06 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | PMP-diketopiperazine adducts form at the active site of a PLP dependent enzyme involved in formycin biosynthesis.
Chem.Commun.(Camb.), 55, 2019
|
|
6SSD
 
 | | Transaminase with PLP bound | | Descriptor: | ForI-PLP, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Naismith, J.H, Gao, S. | | Deposit date: | 2019-09-06 | | Release date: | 2020-01-15 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | PMP-diketopiperazine adducts form at the active site of a PLP dependent enzyme involved in formycin biosynthesis.
Chem.Commun.(Camb.), 55, 2019
|
|
6TM4
 
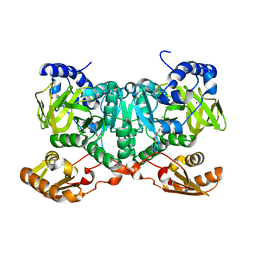 | | NatL2 in complex with two molecules of salicylic acid | | Descriptor: | 2-HYDROXYBENZOIC ACID, ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Naismith, J.H, Song, H. | | Deposit date: | 2019-12-03 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | The Biosynthesis of the Benzoxazole in Nataxazole Proceeds via an Unstable Ester and has Synthetic Utility.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
1A3S
 
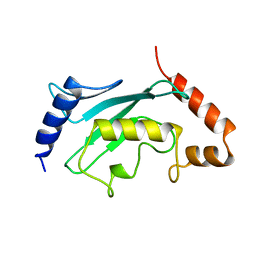 | | HUMAN UBC9 | | Descriptor: | UBC9 | | Authors: | Naismith, J.H, Giraud, M. | | Deposit date: | 1998-01-23 | | Release date: | 1998-05-27 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of ubiquitin-conjugating enzyme 9 displays significant differences with other ubiquitin-conjugating enzymes which may reflect its specificity for sumo rather than ubiquitin.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
2VG0
 
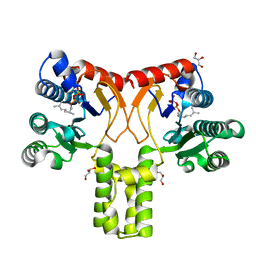 | | Rv1086 citronellyl pyrophosphate complex | | Descriptor: | GERANYL DIPHOSPHATE, GLYCEROL, SHORT-CHAIN Z-ISOPRENYL DIPHOSPHATE SYNTHETASE | | Authors: | Naismith, J.H, Wang, W, Dong, C. | | Deposit date: | 2007-11-07 | | Release date: | 2007-11-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structural basis of chain length control in Rv1086.
J. Mol. Biol., 381, 2008
|
|
5N0O
 
 | |
5N0Q
 
 | |
5N0P
 
 | | Crystal structure of OphA-DeltaC18 in complex with SAH | | Descriptor: | 1,2-ETHANEDIOL, S-ADENOSYL-L-HOMOCYSTEINE, peptide N-methyltranferase | | Authors: | Naismith, J.H, Song, H. | | Deposit date: | 2017-02-03 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | A molecular mechanism for the enzymatic methylation of nitrogen atoms within peptide bonds.
Sci Adv, 4, 2018
|
|
5H8C
 
 | | Truncated XPD | | Descriptor: | IRON/SULFUR CLUSTER, XPD/Rad3 related DNA helicase | | Authors: | Naismith, J.H, Constantinescu, D. | | Deposit date: | 2015-12-23 | | Release date: | 2016-01-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Mechanism of DNA loading by the DNA repair helicase XPD.
Nucleic Acids Res., 44, 2016
|
|
5H8W
 
 | | XPD mechanism | | Descriptor: | ATP-dependent DNA helicase Ta0057, DNA (5'-D(P*TP*AP*CP*GP*A)-3'), IRON/SULFUR CLUSTER, ... | | Authors: | Naismith, J.H, Constantinescu, D. | | Deposit date: | 2015-12-24 | | Release date: | 2016-01-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanism of DNA loading by the DNA repair helicase XPD.
Nucleic Acids Res., 44, 2016
|
|
1TEI
 
 | | STRUCTURE OF CONCANAVALIN A COMPLEXED TO BETA-D-GLCNAC (1,2)ALPHA-D-MAN-(1,6)[BETA-D-GLCNAC(1,2)ALPHA-D-MAN (1,6)]ALPHA-D-MAN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose, CALCIUM ION, CONCANAVALIN A, ... | | Authors: | Naismith, J.H, Moothoo, D.N. | | Deposit date: | 1997-05-28 | | Release date: | 1997-08-20 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Concanavalin A distorts the beta-GlcNAc-(1-->2)-Man linkage of beta-GlcNAc-(1-->2)-alpha-Man-(1-->3)-[beta-GlcNAc-(1-->2)-alpha-Man- (1-->6)]-Man upon binding.
Glycobiology, 8, 1998
|
|
5AJI
 
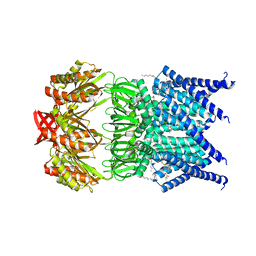 | | MscS D67R1 high resolution | | Descriptor: | DECANE, HEXANE, N-OCTANE, ... | | Authors: | Naismith, J.H, Pliotas, C. | | Deposit date: | 2015-02-24 | | Release date: | 2015-07-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | The Role of Lipids in Mechanosensation.
Nat.Struct.Mol.Biol., 22, 2015
|
|
1QGL
 
 | |
7BIS
 
 | |
7BK9
 
 | |
