8K8F
 
 | | De novo design protein -N7 | | Descriptor: | De novo design protein N7, GLYCEROL | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-07-29 | | Release date: | 2024-07-31 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | De novo protein design with a denoising diffusion network independent of pretrained structure prediction models.
Nat.Methods, 21, 2024
|
|
8K7Z
 
 | | De novo design protein -N1 | | Descriptor: | De novo design protein | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-07-27 | | Release date: | 2024-07-31 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | De novo protein design with a denoising diffusion network independent of pretrained structure prediction models.
Nat.Methods, 21, 2024
|
|
8K7M
 
 | | De novo design protein -T01 | | Descriptor: | De novo design protein | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-07-26 | | Release date: | 2024-07-31 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | De novo protein design with a denoising diffusion network independent of pretrained structure prediction models.
Nat.Methods, 21, 2024
|
|
8K83
 
 | | De novo design protein -N2 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, De novo design protein, GLYCEROL | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-07-28 | | Release date: | 2024-07-31 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | De novo protein design with a denoising diffusion network independent of pretrained structure prediction models.
Nat.Methods, 21, 2024
|
|
8K84
 
 | | De novo design protein -N3 | | Descriptor: | DI(HYDROXYETHYL)ETHER, De novo design protein | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-07-28 | | Release date: | 2024-07-31 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | De novo protein design with a denoising diffusion network independent of pretrained structure prediction models.
Nat.Methods, 21, 2024
|
|
8KAC
 
 | | De novo design protein -NX1 | | Descriptor: | De novo design protein -NX1 | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-08-02 | | Release date: | 2024-08-07 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | De novo protein design with a denoising diffusion network independent of pretrained structure prediction models.
Nat.Methods, 21, 2024
|
|
8K8G
 
 | | De novo design protein -N9 | | Descriptor: | De novo design protein | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-07-29 | | Release date: | 2024-08-07 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | De novo protein design with a denoising diffusion network independent of pretrained structure prediction models.
Nat.Methods, 21, 2024
|
|
8KC4
 
 | | De novo design protein -NA05 | | Descriptor: | De novo design protein -NA05 | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-08-06 | | Release date: | 2024-08-07 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | De novo protein design with a denoising diffusion network independent of pretrained structure prediction models.
Nat.Methods, 21, 2024
|
|
8KDQ
 
 | | De novo design protein -T03 | | Descriptor: | De novo design protein -T03 | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-08-09 | | Release date: | 2024-08-14 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | De novo protein design with a denoising diffusion network independent of pretrained structure prediction models.
Nat.Methods, 21, 2024
|
|
8KC1
 
 | | De novo design protein -NX5 | | Descriptor: | De novo design protein -NX5 | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-08-05 | | Release date: | 2024-08-07 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | De novo protein design with a denoising diffusion network independent of pretrained structure prediction models.
Nat.Methods, 21, 2024
|
|
8KC8
 
 | | De novo design protein -T11 | | Descriptor: | De novo design protein -T11 | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-08-06 | | Release date: | 2024-08-07 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | De novo protein design with a denoising diffusion network independent of pretrained structure prediction models.
Nat.Methods, 21, 2024
|
|
8KCJ
 
 | | De novo design protein -N7 | | Descriptor: | De novo design protein -N7, GLYCEROL | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-08-07 | | Release date: | 2024-08-14 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | De novo protein design with a denoising diffusion network independent of pretrained structure prediction models.
Nat.Methods, 21, 2024
|
|
8KA7
 
 | | De novo design protein -NB7 | | Descriptor: | De novo design protein -NB7 | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-08-02 | | Release date: | 2024-08-07 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | De novo protein design with a denoising diffusion network independent of pretrained structure prediction models.
Nat.Methods, 21, 2024
|
|
5HV2
 
 | |
8KC5
 
 | | De novo design protein -T09 | | Descriptor: | De novo design protein -T09 | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-08-06 | | Release date: | 2024-08-07 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | De novo protein design with a denoising diffusion network independent of pretrained structure prediction models.
Nat.Methods, 21, 2024
|
|
8KA6
 
 | | De novo design protein -NA7 | | Descriptor: | De novo design protein -NA7 | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-08-02 | | Release date: | 2024-08-07 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | De novo protein design with a denoising diffusion network independent of pretrained structure prediction models.
Nat.Methods, 21, 2024
|
|
8KCK
 
 | | De novo design protein -N9 | | Descriptor: | De novo design protein -N9 | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-08-07 | | Release date: | 2024-08-14 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | De novo protein design with a denoising diffusion network independent of pretrained structure prediction models.
Nat.Methods, 21, 2024
|
|
8KC0
 
 | | De novo design protein -NB8 | | Descriptor: | De novo design protein -NB8 | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-08-05 | | Release date: | 2024-08-07 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | De novo protein design with a denoising diffusion network independent of pretrained structure prediction models.
Nat.Methods, 21, 2024
|
|
5GH9
 
 | | Crystal structure of CBP Bromodomain with H3K56ac peptide | | Descriptor: | CREB-binding protein, Histone H3 | | Authors: | Xu, L. | | Deposit date: | 2016-06-19 | | Release date: | 2017-06-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.451 Å) | | Cite: | Structural insight into CBP bromodomain-mediated recognition of acetylated histone H3K56ac
FEBS J., 2017
|
|
5XEX
 
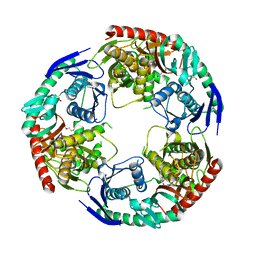 | | Crystal structure of S.aureus PNPase catalytic domain | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, PYROPHOSPHATE, ... | | Authors: | Wang, X, Zhang, X, Zang, J. | | Deposit date: | 2017-04-06 | | Release date: | 2017-10-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Enolase binds to RnpA in competition with PNPase in Staphylococcus aureus
FEBS Lett., 591, 2017
|
|
5JD0
 
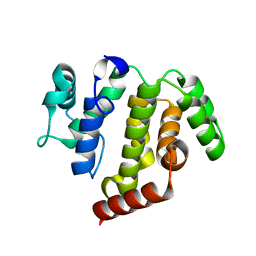 | | crystal structure of ARAP3 RhoGAP domain | | Descriptor: | Arf-GAP with Rho-GAP domain, ANK repeat and PH domain-containing protein 3 | | Authors: | Bao, H, Li, F, Wu, J, Shi, Y. | | Deposit date: | 2016-04-15 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for the Specific Recognition of RhoA by the Dual GTPase-activating Protein ARAP3
J.Biol.Chem., 291, 2016
|
|
8K2S
 
 | |
8K2R
 
 | |
8K2T
 
 | |
5ZVB
 
 | | APOBEC3F Chimeric Catalytic Domain in Complex with DNA(dT9) | | Descriptor: | APEBEC3F/ssDNA-T9, CACODYLATE ION, DNA (5'-D(*AP*TP*TP*TP*TP*CP*AP*AP*T)-3'), ... | | Authors: | Cheng, C, Zhang, T.L, Wang, C.X, Lan, W.X, Ding, J.P, Cao, C.Y. | | Deposit date: | 2018-05-09 | | Release date: | 2018-11-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Cytidine Deaminase Human APOBEC3F Chimeric Catalytic Domain in Complex with DNA
Chin.J.Chem., 36, 2018
|
|
