6MQM
 
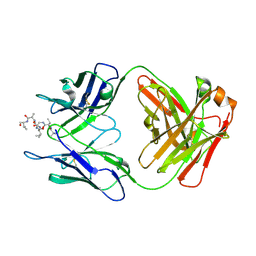 | |
6MQR
 
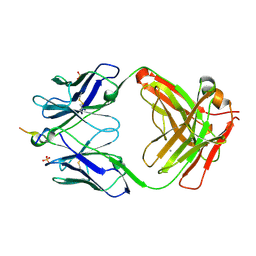 | | Vaccine-elicited NHP FP-targeting neutralizing antibody 0PV-a.01 in complex with FP (residue 512-519) | | Descriptor: | CALCIUM ION, HIV fusion peptide residue 512-519, SULFATE ION, ... | | Authors: | Xu, K, Wang, Y, Kwong, P.D. | | Deposit date: | 2018-10-10 | | Release date: | 2019-07-31 | | Last modified: | 2019-11-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Antibody Lineages with Vaccine-Induced Antigen-Binding Hotspots Develop Broad HIV Neutralization.
Cell, 178, 2019
|
|
6N16
 
 | |
6N1W
 
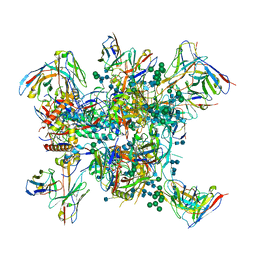 | |
6N1V
 
 | |
6MPG
 
 | | Cryo-EM structure at 3.2 A resolution of HIV-1 fusion peptide-directed antibody, A12V163-b.01, elicited by vaccination of Rhesus macaques, in complex with stabilized HIV-1 Env BG505 DS-SOSIP, which was also bound to antibodies VRC03 and PGT122 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, A12V163-b.01 Heavy Chain, ... | | Authors: | Acharya, P, Kwong, P.D. | | Deposit date: | 2018-10-06 | | Release date: | 2019-07-24 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Antibody Lineages with Vaccine-Induced Antigen-Binding Hotspots Develop Broad HIV Neutralization.
Cell, 178, 2019
|
|
5YU9
 
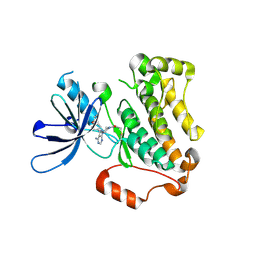 | | Crystal structure of EGFR 696-1022 T790M in complex with Ibrutinib | | Descriptor: | 1-{(3R)-3-[4-amino-3-(4-phenoxyphenyl)-1H-pyrazolo[3,4-d]pyrimidin-1-yl]piperidin-1-yl}prop-2-en-1-one, CHLORIDE ION, Epidermal growth factor receptor | | Authors: | Yan, X.E, Yun, C.H. | | Deposit date: | 2017-11-21 | | Release date: | 2017-12-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Ibrutinib targets mutant-EGFR kinase with a distinct binding conformation.
Oncotarget, 7, 2016
|
|
5VVM
 
 | |
5VT8
 
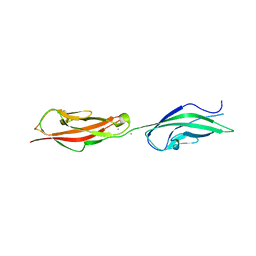 | |
5W1D
 
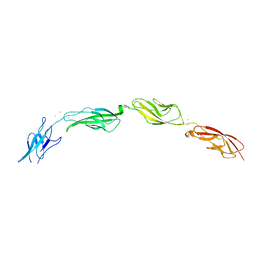 | |
5W4T
 
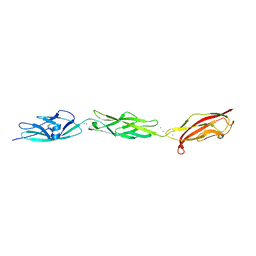 | | Crystal Structure of Fish Cadherin-23 EC1-3 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, ... | | Authors: | De-la-Torre, P, Sotomayor, M. | | Deposit date: | 2017-06-12 | | Release date: | 2018-06-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Zooming in on Cadherin-23: Structural Diversity and Potential Mechanisms of Inherited Deafness.
Structure, 26, 2018
|
|
5WJR
 
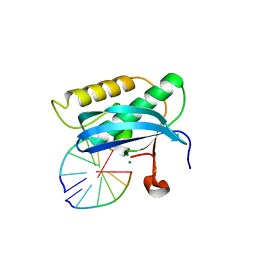 | |
5X39
 
 | |
5X34
 
 | |
5F7E
 
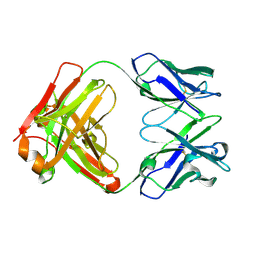 | | Crystal structure of germ-line precursor of 3BNC60 Fab | | Descriptor: | Fab heavy chain, Fab light chain | | Authors: | Sievers, S.A, Scharf, L, Jiang, S, Bjorkman, P.J. | | Deposit date: | 2015-12-08 | | Release date: | 2016-04-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for germline antibody recognition of HIV-1 immunogens.
Elife, 5, 2016
|
|
3GCW
 
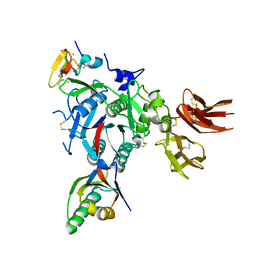 | | PCSK9:EGFA(H306Y) | | Descriptor: | CALCIUM ION, Low-density lipoprotein receptor, Proprotein convertase subtilisin/kexin type 9 | | Authors: | Kwon, H.J. | | Deposit date: | 2009-02-22 | | Release date: | 2009-03-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Antagonism of Secreted PCSK9 Increases Low Density Lipoprotein Receptor Expression in HepG2 Cells.
J.Biol.Chem., 284, 2009
|
|
5FA2
 
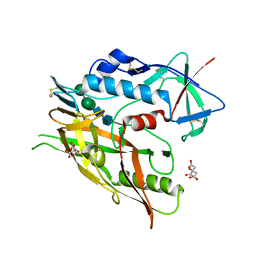 | | Crystal structure of 426c.TM4deltaV1-3 p120 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CITRATE ANION, THIOCYANATE ION, ... | | Authors: | Scharf, L, Bjorkman, P.J. | | Deposit date: | 2015-12-10 | | Release date: | 2016-04-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for germline antibody recognition of HIV-1 immunogens.
Elife, 5, 2016
|
|
5FHF
 
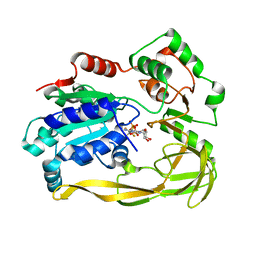 | | Crystal structure of Bacteroides sp Pif1 in complex with ADP-AlF4 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, TETRAFLUOROALUMINATE ION, Uncharacterized protein | | Authors: | Zhou, X, Ren, W, Bharath, S.R, Song, H. | | Deposit date: | 2015-12-22 | | Release date: | 2016-03-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and Functional Insights into the Unwinding Mechanism of Bacteroides sp Pif1
Cell Rep, 14, 2016
|
|
5FHE
 
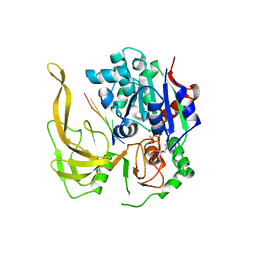 | | Crystal structure of Bacteroides Pif1 bound to ssDNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), MAGNESIUM ION, ... | | Authors: | Zhou, X, Ren, W, Bharath, S.R, Song, H. | | Deposit date: | 2015-12-22 | | Release date: | 2016-03-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and Functional Insights into the Unwinding Mechanism of Bacteroides sp Pif1
Cell Rep, 14, 2016
|
|
5FEC
 
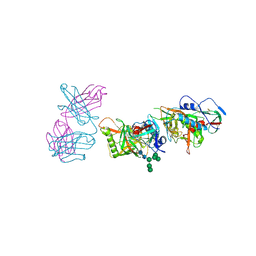 | |
5XMK
 
 | | Cryo-EM structure of the ATP-bound Vps4 mutant-E233Q complex with Vta1 (masked) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Vacuolar protein sorting-associated protein 4, Vacuolar protein sorting-associated protein VTA1 | | Authors: | Sun, S, Li, L, Yang, F, Wang, X, Fan, F, Li, X, Wang, H, Sui, S. | | Deposit date: | 2017-05-15 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.18 Å) | | Cite: | Cryo-EM structures of the ATP-bound Vps4(E233Q) hexamer and its complex with Vta1 at near-atomic resolution
Nat Commun, 8, 2017
|
|
5FHG
 
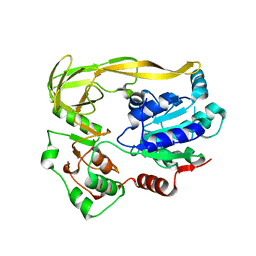 | |
5WJM
 
 | | Crystal Structure of Mouse Cadherin-23 EC17-18 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Cadherin-23, ... | | Authors: | De-la-Torre, P, Sotomayor, M. | | Deposit date: | 2017-07-23 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Zooming in on Cadherin-23: Structural Diversity and Potential Mechanisms of Inherited Deafness.
Structure, 26, 2018
|
|
5XMI
 
 | | Cryo-EM Structure of the ATP-bound VPS4 mutant-E233Q hexamer (masked) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Vacuolar protein sorting-associated protein 4 | | Authors: | Sun, S, Li, L, Yang, F, Wang, X, Fan, F, Li, X, Wang, H, Sui, S. | | Deposit date: | 2017-05-15 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structures of the ATP-bound Vps4(E233Q) hexamer and its complex with Vta1 at near-atomic resolution
Nat Commun, 8, 2017
|
|
5VH2
 
 | |
