1EE6
 
 | | CRYSTAL STRUCTURE OF PECTATE LYASE FROM BACILLUS SP. STRAIN KSM-P15. | | Descriptor: | CALCIUM ION, PECTATE LYASE | | Authors: | Akita, M, Suzuki, A, Kobayashi, T, Ito, S, Yamane, T. | | Deposit date: | 2000-01-31 | | Release date: | 2001-01-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The first structure of pectate lyase belonging to polysaccharide lyase family 3.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1G01
 
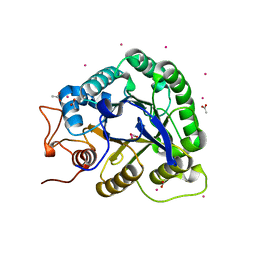 | | ALKALINE CELLULASE K CATALYTIC DOMAIN | | Descriptor: | ACETIC ACID, CADMIUM ION, ENDOGLUCANASE | | Authors: | Shirai, T, Ishida, H, Noda, J, Yamane, T, Ozaki, K, Hakamada, Y, Ito, S. | | Deposit date: | 2000-10-05 | | Release date: | 2001-08-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of alkaline cellulase K: insight into the alkaline adaptation of an industrial enzyme.
J.Mol.Biol., 310, 2001
|
|
1G0C
 
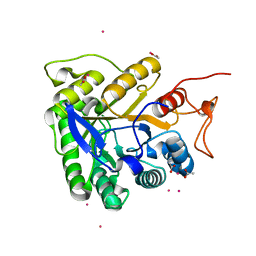 | | ALKALINE CELLULASE K CATALYTIC DOMAIN-CELLOBIOSE COMPLEX | | Descriptor: | ACETIC ACID, CADMIUM ION, ENDOGLUCANASE, ... | | Authors: | Shirai, T, Ishida, H, Noda, J, Yamane, T, Ozaki, K, Hakamada, Y, Ito, S. | | Deposit date: | 2000-10-05 | | Release date: | 2001-08-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of alkaline cellulase K: insight into the alkaline adaptation of an industrial enzyme.
J.Mol.Biol., 310, 2001
|
|
1IS3
 
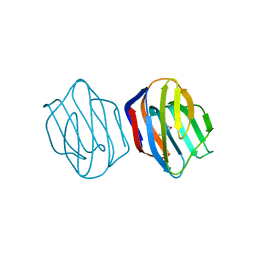 | | LACTOSE AND MES-LIGANDED CONGERIN II | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CONGERIN II, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Shirai, T, Matsui, Y, Shionyu-Mitsuyama, C, Yamane, T, Kamiya, H, Ishii, C, Ogawa, T, Muramoto, K. | | Deposit date: | 2001-11-12 | | Release date: | 2002-09-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of a conger eel galectin (congerin II) at 1.45 A resolution: Implication for the accelerated evolution of a new ligand-binding site following gene duplication
J.MOL.BIOL., 321, 2002
|
|
1IS5
 
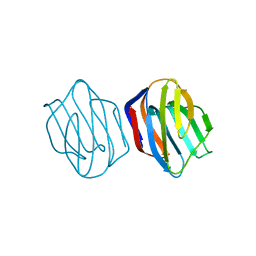 | | Ligand free Congerin II | | Descriptor: | Congerin II | | Authors: | Shirai, T, Matsui, Y, Shionyu-Mitsuyama, C, Yamane, T, Kamiya, H, Ishii, C, Ogawa, T, Muramoto, K. | | Deposit date: | 2001-11-12 | | Release date: | 2002-09-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a conger eel galectin (congerin II) at 1.45 A resolution: Implication for the accelerated evolution of a new ligand-binding site following gene duplication
J.MOL.BIOL., 321, 2002
|
|
1IS6
 
 | | MES-Liganded Congerin II | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Congerin II | | Authors: | Shirai, T, Matsui, Y, Shionyu-Mitsuyama, C, Yamane, T, Kamiya, H, Ishii, C, Ogawa, T, Muramoto, K. | | Deposit date: | 2001-11-12 | | Release date: | 2002-09-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a conger eel galectin (congerin II) at 1.45 A resolution: Implication for the accelerated evolution of a new ligand-binding site following gene duplication
J.MOL.BIOL., 321, 2002
|
|
1IS4
 
 | | LACTOSE-LIGANDED CONGERIN II | | Descriptor: | CONGERIN II, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Shirai, T, Matsui, Y, Shionyu-Mitsuyama, C, Yamane, T, Kamiya, H, Ishii, C, Ogawa, T, Muramoto, K. | | Deposit date: | 2001-11-12 | | Release date: | 2002-09-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of a Conger Eel Galectin (Congerin II) at 1.45 A Resolution: Implication for the Accelerated Evolution of a New Ligand-Binding Site Following Gene Duplication
J.Mol.Biol., 321, 2002
|
|
1VDP
 
 | | The crystal structure of the monoclinic form of hen egg white lysozyme at 1.7 angstroms resolution in space | | Descriptor: | Lysozyme C | | Authors: | Aibara, S, Suzuki, A, Kidera, A, Shibata, K, Yamane, T, DeLucas, L.J, Hirose, M. | | Deposit date: | 2004-03-24 | | Release date: | 2004-04-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of the monoclinic form of hen egg white lysozyme at 1.7 angstroms resolution in space
to be published
|
|
1VDQ
 
 | | The crystal structure of the orthorhombic form of hen egg white lysozyme at 1.5 angstroms resolution | | Descriptor: | Lysozyme C | | Authors: | Aibara, S, Suzuki, A, Kidera, A, Shibata, K, Yamane, T, DeLucas, L.J, Hirose, M. | | Deposit date: | 2004-03-24 | | Release date: | 2004-04-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of the orthorhombic form of hen egg white lysozyme at 1.5 angstroms resolution
to be published
|
|
1VDT
 
 | | The crystal structure of the tetragonal form of hen egg white lysozyme at 1.7 angstroms resolution under basic conditions in space | | Descriptor: | Lysozyme C | | Authors: | Aibara, S, Suzuki, A, Kidera, A, Shibata, K, Yamane, T, DeLucas, L.J, Hirose, M. | | Deposit date: | 2004-03-24 | | Release date: | 2004-04-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of the tetragonal form of hen egg white lysozyme at 1.7 angstroms resolution under basic conditions in space
to be published
|
|
1VED
 
 | | The crystal structure of the orthorhombic form of hen egg white lysozyme at 1.9 angstroms resolution in space | | Descriptor: | Lysozyme C | | Authors: | Aibara, S, Suzuki, A, Kidera, A, Shibata, K, Yamane, T, DeLucas, L.J, Hirose, M. | | Deposit date: | 2004-03-30 | | Release date: | 2004-04-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of the orthorhombic form of hen egg white lysozyme at 1.9 angstroms resolution in space
To be Published
|
|
1VDS
 
 | | The crystal structure of the tetragonal form of hen egg white lysozyme at 1.6 angstroms resolution in space | | Descriptor: | Lysozyme C | | Authors: | Aibara, S, Suzuki, A, Kidera, A, Shibata, K, Yamane, T, DeLucas, L.J, Hirose, M. | | Deposit date: | 2004-03-24 | | Release date: | 2004-04-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of the tetragonal form of hen egg white lysozyme at 1.6 angstroms resolution in space
to be published
|
|
3RKS
 
 | | Crystal Structure of the Manihot esculenta Hydroxynitrile Lyase (MeHNL) K176P mutant | | Descriptor: | GLYCEROL, Hydroxynitrilase | | Authors: | Cielo, C.B.C, Yamane, T, Asano, Y, Dadashipour, M, Suzuki, A, Mizushima, T, Komeda, H. | | Deposit date: | 2011-04-18 | | Release date: | 2012-06-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystallographic Studies of Manihot esculenta hydroxynitrile lyase Lysine-to-Proline mutants
To be Published
|
|
2GGQ
 
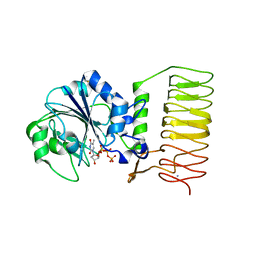 | | complex of hypothetical glucose-1-phosphate thymidylyltransferase from sulfolobus tokodaii | | Descriptor: | 401aa long hypothetical glucose-1-phosphate thymidylyltransferase, IODIDE ION, THYMIDINE-5'-TRIPHOSPHATE | | Authors: | Rajakannan, V, Mizushima, T, Suzuki, A, Masui, R, Kuramitsu, S, Yamane, T. | | Deposit date: | 2006-03-24 | | Release date: | 2007-03-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | complex of hypothetical glucose-1-phosphate thymidylyltransferase from sulfolobus tokodaii
To be published
|
|
2GGO
 
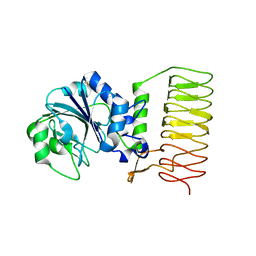 | | Crystal Structure of glucose-1-phosphate thymidylyltransferase from Sulfolobus tokodaii | | Descriptor: | 401aa long hypothetical glucose-1-phosphate thymidylyltransferase | | Authors: | Rajakannan, V, Mizushima, T, Suzuki, A, Masui, R, Kuramitsu, S, Yamane, T. | | Deposit date: | 2006-03-24 | | Release date: | 2007-03-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of glucose-1-phosphate thymidylyltransferase from
Sulfolobus tokodaii
To be published
|
|
1WSD
 
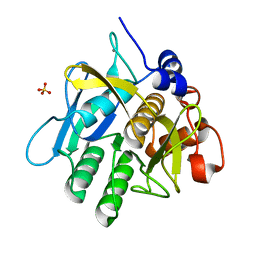 | | Alkaline M-protease form I crystal structure | | Descriptor: | CALCIUM ION, M-protease, SULFATE ION | | Authors: | Shirai, T, Suzuki, A, Yamane, T, Ashida, T, Kobayashi, T, Hitomi, J, Ito, S. | | Deposit date: | 2004-11-05 | | Release date: | 2004-11-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High-resolution crystal structure of M-protease: phylogeny aided analysis of the high-alkaline adaptation mechanism
Protein Eng., 10, 1997
|
|
1V9Q
 
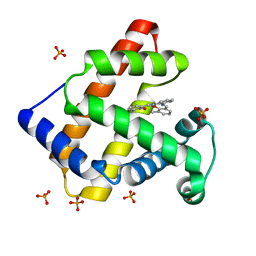 | | Crystal Structure of an Artificial Metalloprotein:Mn(III)(3,3'-Me2-salophen)/apo-A71G Myoglobin | | Descriptor: | 'N,N'-BIS-(2-HYDROXY-3-METHYL-BENZYLIDENE)-BENZENE-1,2-DIAMINE', MANGANESE (III) ION, Myoglobin, ... | | Authors: | Ueno, T, Koshiyama, T, Kono, M, Kondo, K, Ohashi, M, Suzuki, A, Yamane, T, Watanabe, Y. | | Deposit date: | 2004-01-29 | | Release date: | 2005-05-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Coordinated Design of Cofactor and Active Site Structures in Development of New Protein Catalysts
J.Am.Chem.Soc., 127, 2005
|
|
2DCN
 
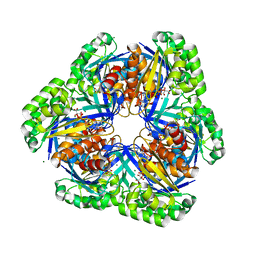 | | Crystal structure of 2-keto-3-deoxygluconate kinase from Sulfolobus tokodaii complexed with 2-keto-6-phosphogluconate (alpha-furanose form) | | Descriptor: | 6-O-phosphono-beta-D-psicofuranosonic acid, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Okazaki, S, Onda, H, Suzuki, A, Kuramitsu, S, Masui, R, Yamane, T. | | Deposit date: | 2006-01-10 | | Release date: | 2006-01-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of 2-keto-3-deoxygluconate kinase from Sulfolobus tokodaii complexed with 2-keto-6-phosphogluconate
To be Published
|
|
2E31
 
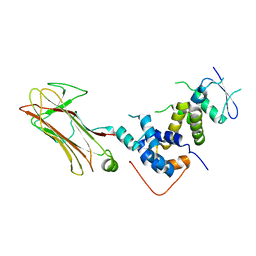 | | Structural basis for selection of glycosylated substrate by SCFFbs1 ubiquitin ligase | | Descriptor: | F-box only protein 2, S-phase kinase-associated protein 1A | | Authors: | Mizushima, T, Yoshida, Y, Kumanomidou, T, Hasegawa, Y, Yamane, T, Tanaka, K. | | Deposit date: | 2006-11-20 | | Release date: | 2007-03-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the selection of glycosylated substrates by SCFFbs1 ubiquitin ligase
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2EB8
 
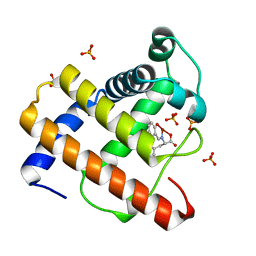 | | Crystal Structure of Cu(II)(Sal-Phe)/apo-Myoglobin | | Descriptor: | (N-SALICYLIDEN-L-PHENYLALANATO)-COPPER(II), Myoglobin, PHOSPHATE ION | | Authors: | Abe, S, Okazaki, S, Ueno, T, Hikage, T, Suzuki, A, Yamane, T, Watanabe, Y. | | Deposit date: | 2007-02-07 | | Release date: | 2007-07-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Design and Structure Analysis of Artificial Metalloproteins: Selective Coordination of His64 to Copper Complexes with Square-Planar Structure in the apo-Myoglobin Scaffold
Inorg.Chem., 46, 2007
|
|
2EF2
 
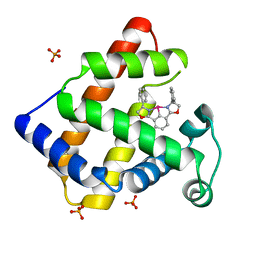 | | Crystal Structure of an Artificial Metalloprotein:Rh(Phebox-Ph)/apo-A71G Myoglobin | | Descriptor: | Myoglobin, PHOSPHATE ION, [2,6-BIS(4-PHENYL)-1,3-OXAZOLIN-2-YL]RHODIUM(III) | | Authors: | Abe, S, Satake, Y, Okazaki, S, Ueno, T, Hikage, T, Suzuki, A, Yamane, T, Nakajima, H, Watanabe, Y. | | Deposit date: | 2007-02-20 | | Release date: | 2007-10-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Incorporation of a Phebox Rhodium Complex into apo-Myoglobin Affords a Stable Organometallic Protein Showing Unprecedented Arrangement of the Complex in the Cavity
ORGANOMETALLICS, 26, 2007
|
|
2E32
 
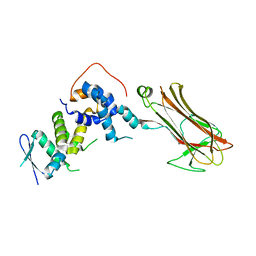 | | Structural basis for selection of glycosylated substrate by SCFFbs1 ubiquitin ligase | | Descriptor: | F-box only protein 2, S-phase kinase-associated protein 1A | | Authors: | Mizushima, T, Yoshida, Y, Kumanomidou, T, Hasegawa, Y, Yamane, T, Tanaka, K. | | Deposit date: | 2006-11-20 | | Release date: | 2007-03-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.52 Å) | | Cite: | Structural basis for the selection of glycosylated substrates by SCFFbs1 ubiquitin ligase
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2EB9
 
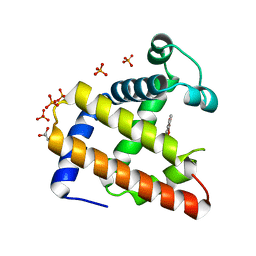 | | Crystal Structure of Cu(II)(Sal-Leu)/apo-Myoglobin | | Descriptor: | (N-SALICYLIDEN-L-LEUCINATO)-COPPER(II), GLYCEROL, Myoglobin, ... | | Authors: | Abe, S, Okazaki, S, Ueno, T, Hikage, T, Suzuki, A, Yamane, T, Watanabe, Y. | | Deposit date: | 2007-02-07 | | Release date: | 2007-07-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design and Structure Analysis of Artificial Metalloproteins: Selective Coordination of His64 to Copper Complexes with Square-Planar Structure in the apo-Myoglobin Scaffold
Inorg.Chem., 46, 2007
|
|
2E33
 
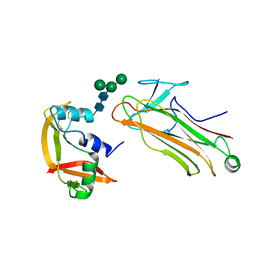 | | Structural basis for selection of glycosylated substrate by SCFFbs1 ubiquitin ligase | | Descriptor: | F-box only protein 2, Ribonuclease pancreatic, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Mizushima, T, Yoshida, Y, Kumanomidou, T, Hasegawa, Y, Yamane, T, Tanaka, K. | | Deposit date: | 2006-11-20 | | Release date: | 2007-03-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for the selection of glycosylated substrates by SCFFbs1 ubiquitin ligase
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
1X23
 
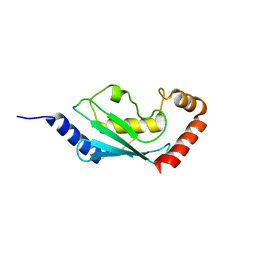 | | Crystal structure of ubch5c | | Descriptor: | Ubiquitin-conjugating enzyme E2 D3 | | Authors: | Nakanishi, M, Teshima, N, Mizushima, T, Murata, S, Tanaka, K, Yamane, T. | | Deposit date: | 2005-04-19 | | Release date: | 2005-05-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of ubch5c
To be Published
|
|
