8FNT
 
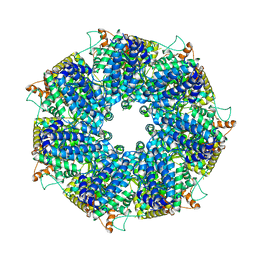 | | Structure of RdrA from Escherichia coli RADAR defense system | | Descriptor: | Archaeal ATPase | | Authors: | Duncan-Lowey, B, Johnson, A.G, Rawson, S, Mayer, M.L, Kranzusch, P.J. | | Deposit date: | 2022-12-28 | | Release date: | 2023-02-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Cryo-EM structure of the RADAR supramolecular anti-phage defense complex.
Cell, 186, 2023
|
|
8FNV
 
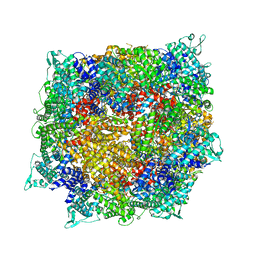 | | Structure of RdrB from Escherichia coli RADAR defense system | | Descriptor: | Adenosine deaminase, ZINC ION | | Authors: | Duncan-Lowey, B, Johnson, A.G, Rawson, S, Mayer, M.L, Kranzusch, P.J. | | Deposit date: | 2022-12-28 | | Release date: | 2023-02-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.11 Å) | | Cite: | Cryo-EM structure of the RADAR supramolecular anti-phage defense complex.
Cell, 186, 2023
|
|
8FNU
 
 | | Structure of RdrA from Streptococcus suis RADAR defense system | | Descriptor: | KAP NTPase domain-containing protein | | Authors: | Duncan-Lowey, B, Johnson, A.G, Rawson, S, Mayer, M.L, Kranzusch, P.J. | | Deposit date: | 2022-12-28 | | Release date: | 2023-02-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Cryo-EM structure of the RADAR supramolecular anti-phage defense complex.
Cell, 186, 2023
|
|
8BTO
 
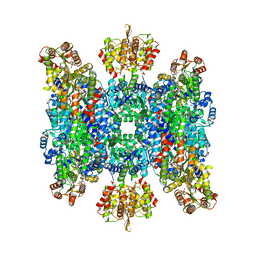 | | Helical structure of BcThsA in complex with 1''-3'gcADPR | | Descriptor: | (2R,3R,3aS,5S,6R,7S,8R,11R,13S,15aR)-2-(6-amino-9H-purin-9-yl)-3,6,7,11,13-pentahydroxyoctahydro-2H,5H,11H,13H-5,8-epoxy-11lambda~5~,13lambda~5~-furo[2,3-g][1,3,5,9,2,4]tetraoxadiphosphacyclotetradecine-11,13-dione, NAD(+) hydrolase ThsA, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Tamulaitiene, G, Sasnauskas, G, Sabonis, D. | | Deposit date: | 2022-11-29 | | Release date: | 2024-02-21 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Activation of Thoeris antiviral system via SIR2 effector filament assembly.
Nature, 627, 2024
|
|
8BTN
 
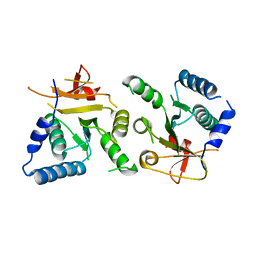 | | Crystal structure of BcThsB | | Descriptor: | TIR domain-containing protein | | Authors: | Tamulaitiene, G, Sabonis, D. | | Deposit date: | 2022-11-29 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Activation of Thoeris antiviral system via SIR2 effector filament assembly.
Nature, 627, 2024
|
|
8BTP
 
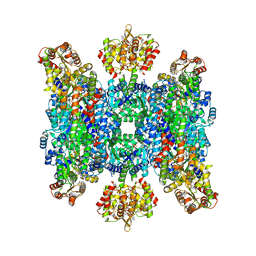 | | Helical structure of BcThsA in complex with 1''-3'gc(etheno)ADPR | | Descriptor: | (1S,3S,4R,5R,7R,15R,16S,17R)-5-imidazo[2,1-f]purin-3-yl-10,12-bis(oxidanyl)-10,12-bis(oxidanylidene)-2,6,9,11,13,18-hexaoxa-10$l^{5},12$l^{5}-diphosphatricyclo[13.2.1.0^{3,7}]octadecane-4,16,17-triol, ETHENO-NAD, NAD(+) hydrolase ThsA | | Authors: | Tamulaitiene, G, Sasnauskas, G, Sabonis, D. | | Deposit date: | 2022-11-29 | | Release date: | 2024-02-21 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Activation of Thoeris antiviral system via SIR2 effector filament assembly.
Nature, 627, 2024
|
|
8SS1
 
 | |
8SRZ
 
 | |
8SM3
 
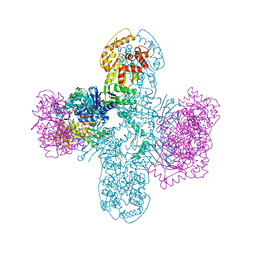 | | Structure of Bacillus cereus VD045 Gabija GajA-GajB Complex | | Descriptor: | Endonuclease GajA, Gabija protein GajB, SULFATE ION | | Authors: | Antine, S.P, Mooney, S.E, Johnson, A.G, Kranzusch, P.J. | | Deposit date: | 2023-04-25 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of Gabija anti-phage defence and viral immune evasion.
Nature, 625, 2024
|
|
8TTO
 
 | |
