6SMF
 
 | |
6SME
 
 | |
6YX8
 
 | | The structure of allophycocyanin from cyanobacterium Nostoc sp. WR13, the C2221 crystal form. | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, ... | | Authors: | Patel, H.M, Roszak, A.W, Madamwar, D, Cogdell, R.J. | | Deposit date: | 2020-04-30 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.831 Å) | | Cite: | The high resolution structure of allophycocyanin from cyanobacterium Nostoc sp. WR13
To Be Published
|
|
6YX7
 
 | | The high resolution structure of allophycocyanin from cyanobacterium Nostoc sp. WR13, the P21212 crystal form. | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, ... | | Authors: | Patel, H.M, Roszak, A.W, Madamwar, D, Cogdell, R.J. | | Deposit date: | 2020-04-30 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.419 Å) | | Cite: | The high resolution structure of allophycocyanin from cyanobacterium Nostoc sp. WR13
To Be Published
|
|
6XWK
 
 | | Crystal structure of Phormidium rubidum phycocyanin | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, IMIDAZOLE, ... | | Authors: | Sonani, R.R, Roszak, A.W, Cogdell, R.J, Madamwar, D, Liu, H, Gross, M.L, Blankenship, R.E. | | Deposit date: | 2020-01-23 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Revisiting high-resolution crystal structure of Phormidium rubidum phycocyanin.
Photosyn. Res., 144, 2020
|
|
1BK4
 
 | | CRYSTAL STRUCTURE OF RABBIT LIVER FRUCTOSE-1,6-BISPHOSPHATASE AT 2.3 ANGSTROM RESOLUTION | | Descriptor: | MAGNESIUM ION, PROTEIN (FRUCTOSE-1,6-BISPHOSPHATASE), SULFATE ION | | Authors: | Ghosh, D, Weeks, C.M, Erman, M, Roszak, A.W, Kaiser, R, Jornvall, H. | | Deposit date: | 1998-07-14 | | Release date: | 1998-07-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of rabbit liver fructose 1,6-bisphosphatase at 2.3 A resolution.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1ZYE
 
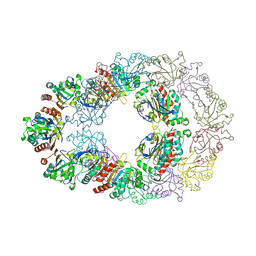 | | Crystal structure analysis of Bovine Mitochondrial Peroxiredoxin III | | Descriptor: | Thioredoxin-dependent peroxide reductase | | Authors: | Cao, Z, Roszak, A.W, Gourlay, L.J, Lindsay, J.G, Isaacs, N.W. | | Deposit date: | 2005-06-10 | | Release date: | 2005-09-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Bovine Mitochondrial Peroxiredoxin III Forms a Two-Ring Catenane
Structure, 13, 2005
|
|
3I4D
 
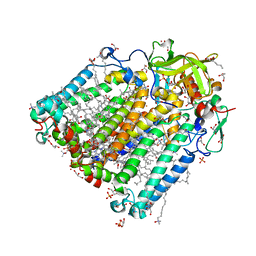 | | Photosynthetic reaction center from rhodobacter sphaeroides 2.4.1 | | Descriptor: | (2R,3S)-heptane-1,2,3-triol, 1,4-DIETHYLENE DIOXIDE, BACTERIOCHLOROPHYLL A, ... | | Authors: | Fujii, R, Adachi, S, Roszak, A.W, Gardiner, A.T, Cogdell, R.J, Isaacs, N.W, Koshihara, S, Hashimoto, H. | | Deposit date: | 2009-07-01 | | Release date: | 2010-12-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure of the carotenoid bound to the reaction centre from Rhodobacter sphaeroides 2.4.1 revealed by time-resolved X-ray crystallography
To be Published
|
|
6HS9
 
 | |
6HSA
 
 | |
6HS8
 
 | |
6HSR
 
 | |
6HSB
 
 | |
6HSU
 
 | |
6HSQ
 
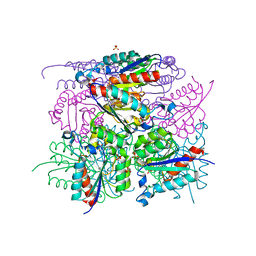 | |
4QKO
 
 | | The Crystal Structure of the Pyocin S2 Nuclease Domain, Immunity Protein Complex at 1.8 Angstroms | | Descriptor: | BROMIDE ION, MAGNESIUM ION, Pyocin-S2, ... | | Authors: | Grinter, R, Josts, I, Roszak, A.W, Cogdell, C.J, Walker, D. | | Deposit date: | 2014-06-07 | | Release date: | 2015-06-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Insights into pyocin S2
To be Published
|
|
4RTD
 
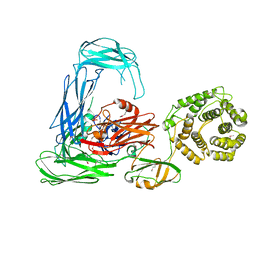 | | Escherichia coli alpha-2-macroglobulin activated by porcine elastase | | Descriptor: | Uncharacterized lipoprotein YfhM | | Authors: | Fyfe, C.D, Grinter, R, Roszak, A.W, Josts, I, Cogdell, R.J, Walker, D. | | Deposit date: | 2014-11-14 | | Release date: | 2015-07-15 | | Last modified: | 2015-07-29 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Structure of protease-cleaved Escherichia coli alpha-2-macroglobulin reveals a putative mechanism of conformational activation for protease entrapment.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5NB3
 
 | | High resolution C-phycoerythrin from marine cyanobacterium Phormidium sp. A09DM at pH 7.5 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, ... | | Authors: | Sonani, R.R, Roszak, A.W, Ortmann de Percin Northumberland, C, Madamwar, D, Cogdell, R.J. | | Deposit date: | 2017-03-01 | | Release date: | 2017-09-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | An improved crystal structure of C-phycoerythrin from the marine cyanobacterium Phormidium sp. A09DM.
Photosyn. Res., 135, 2018
|
|
5NB4
 
 | | Atomic resolution structure of C-phycoerythrin from marine cyanobacterium Phormidium sp. A09DM at pH 7.5 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, HYDROGENPHOSPHATE ION, ... | | Authors: | Sonani, R.R, Roszak, A.W, Ortmann de Percin Northumberland, C, Madamwar, D, Cogdell, R.J. | | Deposit date: | 2017-03-01 | | Release date: | 2017-09-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | An improved crystal structure of C-phycoerythrin from the marine cyanobacterium Phormidium sp. A09DM.
Photosyn. Res., 135, 2018
|
|
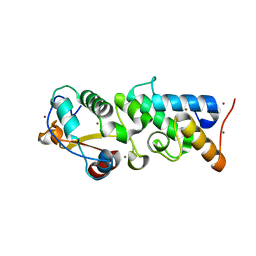
4ZHO
 
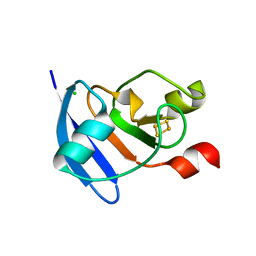 | | The crystal structure of Arabidopsis ferredoxin 2 with 2Fe-2S cluster | | Descriptor: | CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, Ferredoxin-2, ... | | Authors: | Grinter, R, Josts, I, Roszak, A.W, Cogdell, R.J, Walker, D. | | Deposit date: | 2015-04-26 | | Release date: | 2016-08-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structure of the bacterial plant-ferredoxin receptor FusA.
Nat Commun, 7, 2016
|
|
4ZHP
 
 | | The crystal structure of Potato ferredoxin I with 2Fe-2S cluster | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Potato Ferredoxin I | | Authors: | Grinter, R, Josts, I, Roszak, A.W, Cogdell, R.J, Walker, D. | | Deposit date: | 2015-04-26 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structure of the bacterial plant-ferredoxin receptor FusA.
Nat Commun, 7, 2016
|
|
4ZGV
 
 | | The Crystal Structure of the Ferredoxin Receptor FusA from Pectobacterium atrosepticum SCRI1043 | | Descriptor: | Ferredoxin receptor, LAURYL DIMETHYLAMINE-N-OXIDE, octyl beta-D-glucopyranoside | | Authors: | Grinter, R, Josts, I, Roszak, A.W, Cogdell, R.J, Walker, D. | | Deposit date: | 2015-04-24 | | Release date: | 2016-08-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of the bacterial plant-ferredoxin receptor FusA.
Nat Commun, 7, 2016
|
|
5EL8
 
 | |
5ELA
 
 | |
5ELG
 
 | |
