3DXW
 
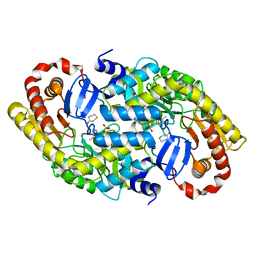 | | The crystal structure of alpha-amino-epsilon-caprolactam racemase from Achromobacter obae complexed with epsilon caprolactam | | Descriptor: | Alpha-amino-epsilon-caprolactam racemase, PYRIDOXAL-5'-PHOSPHATE, azepan-2-one | | Authors: | Okazaki, S, Suzuki, A, Komeda, H, Asano, Y, Yamane, T. | | Deposit date: | 2008-07-25 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | The novel structure of a pyridoxal 5'-phosphate-dependent fold-type I racemase, alpha-amino-epsilon-caprolactam racemase from Achromobacter obae
Biochemistry, 48, 2009
|
|
1IZ1
 
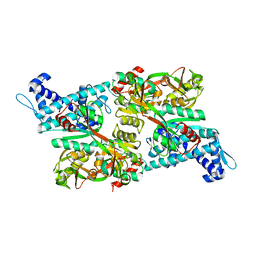 | | CRYSTAL STRUCTURE OF CBNR, A LYSR FAMILY TRANSCRIPTIONAL REGULATOR | | Descriptor: | LysR-type regulatory protein | | Authors: | Muraoka, S, Okumura, R, Ogawa, N, Miyashita, K, Senda, T. | | Deposit date: | 2002-09-18 | | Release date: | 2003-05-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of a Full-length LysR-type Transcriptional Regulator, CbnR: Unusual Combination of Two Subunit Forms and Molecular Bases for Causing and Changing DNA Bend
J.Mol.Biol., 328, 2003
|
|
4FHL
 
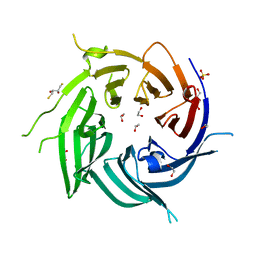 | | Nucleoporin Nup37 from Schizosaccharomyces pombe | | Descriptor: | 1,2-ETHANEDIOL, 1,4-BUTANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ... | | Authors: | Bilokapic, S, Schwartz, T.U. | | Deposit date: | 2012-06-06 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular basis for Nup37 and ELY5/ELYS recruitment to the nuclear pore complex.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4FHN
 
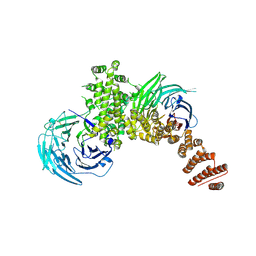 | |
4FCC
 
 | | Glutamate dehydrogenase from E. coli | | Descriptor: | Glutamate dehydrogenase | | Authors: | Bilokapic, S, Schwartz, T.U. | | Deposit date: | 2012-05-24 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis for Nup37 and ELY5/ELYS recruitment to the nuclear pore complex.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4FHM
 
 | |
2L8D
 
 | | Structure/function of the LBR Tudor domain | | Descriptor: | Lamin-B receptor | | Authors: | Liokatis, S, Edlich, C, Soupsana, K, Giannios, I, Sattler, M, Georgatos, S.D, Politou, A.S. | | Deposit date: | 2011-01-10 | | Release date: | 2011-11-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure and molecular interactions of lamin B receptor tudor domain.
J.Biol.Chem., 287, 2012
|
|
4I0O
 
 | |
1IXC
 
 | | Crystal structure of CbnR, a LysR family transcriptional regulator | | Descriptor: | LysR-type regulatory protein | | Authors: | Muraoka, S, Okumura, R, Ogawa, N, Miyashita, K, Senda, T. | | Deposit date: | 2002-06-18 | | Release date: | 2003-06-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of a Full-length LysR-type Transcriptional Regulator, CbnR: Unusual Combination of Two Subunit Forms and Molecular Bases for Causing and Changing DNA Bend
J.Mol.Biol., 328, 2003
|
|
3A4I
 
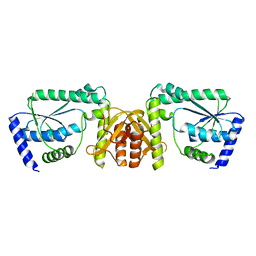 | | Crystal structure of GMP synthetase PH1347 from Pyrococcus horikoshii OT3 | | Descriptor: | GMP synthase [glutamine-hydrolyzing] subunit B | | Authors: | Maruoka, S, Horita, S, Lee, W.C, Nagata, K, Tanokura, M. | | Deposit date: | 2009-07-07 | | Release date: | 2009-07-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal structure of the ATPPase subunit and its substrate-dependent association with the GATase Subunit: a novel regulatory mechanism for a two-subunit-type GMP synthetase from Pyrococcus horikoshii OT3.
J.Mol.Biol., 395, 2010
|
|
2DCN
 
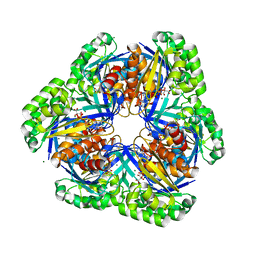 | | Crystal structure of 2-keto-3-deoxygluconate kinase from Sulfolobus tokodaii complexed with 2-keto-6-phosphogluconate (alpha-furanose form) | | Descriptor: | 6-O-phosphono-beta-D-psicofuranosonic acid, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Okazaki, S, Onda, H, Suzuki, A, Kuramitsu, S, Masui, R, Yamane, T. | | Deposit date: | 2006-01-10 | | Release date: | 2006-01-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of 2-keto-3-deoxygluconate kinase from Sulfolobus tokodaii complexed with 2-keto-6-phosphogluconate
To be Published
|
|
2DRW
 
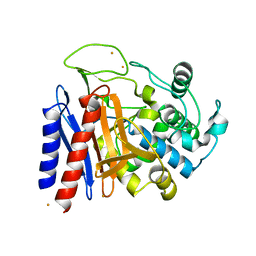 | | The crystal structutre of D-amino acid amidase from Ochrobactrum anthropi SV3 | | Descriptor: | BARIUM ION, D-Amino acid amidase | | Authors: | Okazaki, S, Suzuki, A, Komeda, H, Asano, Y, Yamane, T. | | Deposit date: | 2006-06-15 | | Release date: | 2006-07-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure and Functional Characterization of a D-Stereospecific Amino Acid Amidase from Ochrobactrum anthropi SV3, a New Member of the Penicillin-recognizing Proteins
J.Mol.Biol., 368, 2007
|
|
6KT1
 
 | |
2DNS
 
 | | The crystal structure of D-amino acid amidase from Ochrobactrum anthropi SV3 complexed with D-Phenylalanine | | Descriptor: | BARIUM ION, D-PHENYLALANINE, D-amino acid amidase | | Authors: | Okazaki, S, Suzuki, A, Komeda, H, Asano, Y, Yamane, T. | | Deposit date: | 2006-04-26 | | Release date: | 2006-05-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure and Functional Characterization of a D-Stereospecific Amino Acid Amidase from Ochrobactrum anthropi SV3, a New Member of the Penicillin-recognizing Proteins
J.Mol.Biol., 368, 2007
|
|
2ZUK
 
 | | The crystal structure of alpha-amino-epsilon-caprolactam racemase from Achromobacter obae complexed with epsilon caprolactam (different binding mode) | | Descriptor: | Alpha-amino-epsilon-caprolactam racemase, PYRIDOXAL-5'-PHOSPHATE, azepan-2-one | | Authors: | Okazaki, S, Suzuki, A, Komeda, H, Asano, Y, Yamane, T. | | Deposit date: | 2008-10-18 | | Release date: | 2009-02-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | The novel structure of a pyridoxal 5'-phosphate-dependent fold-type I racemase, alpha-amino-epsilon-caprolactam racemase from Achromobacter obae
Biochemistry, 48, 2009
|
|
2CJB
 
 | | Crystal structure of Methanosarcina barkeri seryl-tRNA synthetase complexed with serine | | Descriptor: | CHLORIDE ION, SERINE, SERYL-TRNA SYNTHETASE, ... | | Authors: | Bilokapic, S, Maier, T, Ahel, D, Gruic-Sovulj, I, Soll, D, Weygand-Durasevic, I, Ban, N. | | Deposit date: | 2006-03-30 | | Release date: | 2006-06-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the Unusual Seryl-tRNA Synthetase Reveals a Distinct Zinc-Dependent Mode of Substrate Recognition
Embo J., 25, 2006
|
|
2CJ9
 
 | | Crystal structure of Methanosarcina barkeri seryl-tRNA synthetase complexed with an analog of seryladenylate | | Descriptor: | 5'-O-(N-(L-SERYL)-SULFAMOYL)ADENOSINE, CHLORIDE ION, SERYL-TRNA SYNTHETASE, ... | | Authors: | Bilokapic, S, Maier, T, Ahel, D, Gruic-Sovulj, I, Soll, D, Weygand-Durasevic, I, Ban, N. | | Deposit date: | 2006-03-29 | | Release date: | 2006-06-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Unusual Seryl-tRNA Synthetase Reveals a Distinct Zinc-Dependent Mode of Substrate Recognition
Embo J., 25, 2006
|
|
2EFX
 
 | | The crystal structure of D-amino acid amidase from Ochrobactrum anthropi SV3 complexed with L-phenylalanine amide | | Descriptor: | BARIUM ION, D-amino acid amidase, PHENYLALANINE AMIDE | | Authors: | Okazaki, S, Suzuki, A, Mizushima, T, Komeda, H, Asano, Y, Yamane, T. | | Deposit date: | 2007-02-26 | | Release date: | 2007-03-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of D-amino-acid amidase complexed with L-phenylalanine and with L-phenylalanine amide: insight into the D-stereospecificity of D-amino-acid amidase from Ochrobactrum anthropi SV3.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
2EFU
 
 | | The crystal structure of D-amino acid amidase from Ochrobactrum anthropi SV3 complexed with L-phenylalanine | | Descriptor: | BARIUM ION, D-Amino acid amidase, PHENYLALANINE | | Authors: | Okazaki, S, Suzuki, A, Mizushima, T, Komeda, H, Asano, Y, Yamane, T. | | Deposit date: | 2007-02-26 | | Release date: | 2007-03-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of D-amino-acid amidase complexed with L-phenylalanine and with L-phenylalanine amide: insight into the D-stereospecificity of D-amino-acid amidase from Ochrobactrum anthropi SV3.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
2CJA
 
 | | Crystal structure of Methanosarcina barkeri seryl-tRNA synthetase complexed with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Bilokapic, S, Maier, T, Ahel, D, Gruic-Sovulj, I, Soll, D, Weygand-Durasevic, I, Ban, N. | | Deposit date: | 2006-03-30 | | Release date: | 2006-06-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the Unusual Seryl-tRNA Synthetase Reveals a Distinct Zinc-Dependent Mode of Substrate Recognition
Embo J., 25, 2006
|
|
2CIM
 
 | | Crystal structure of Methanosarcina barkeri seryl-tRNA synthetase | | Descriptor: | CHLORIDE ION, PLATINUM (II) ION, SERYL-TRNA SYNTHETASE, ... | | Authors: | Bilokapic, S, Maier, T, Ahel, D, Gruic-Sovulj, I, Soll, D, Weygand-Durasevic, I, Ban, N. | | Deposit date: | 2006-03-24 | | Release date: | 2006-06-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structure of the Unusual Seryl-tRNA Synthetase Reveals a Distinct Zinc-Dependent Mode of Substrate Recognition
Embo J., 25, 2006
|
|
2D3W
 
 | | Crystal Structure of Escherichia coli SufC, an ATPase compenent of the SUF iron-sulfur cluster assembly machinery | | Descriptor: | Probable ATP-dependent transporter sufC | | Authors: | Kitaoka, S, Wada, K, Hasegawa, Y, Minami, Y, Takahashi, Y, Fukuyama, K. | | Deposit date: | 2005-10-03 | | Release date: | 2006-01-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Escherichia coli SufC, an ABC-type ATPase component of the SUF iron-sulfur cluster assembly machinery
Febs Lett., 580, 2006
|
|
2D7J
 
 | | Crystal Structure Analysis of Glutamine Amidotransferase from Pyrococcus horikoshii OT3 | | Descriptor: | GMP synthase [glutamine-hydrolyzing] subunit A | | Authors: | Maruoka, S, Lee, W.C, Kamo, M, Kudo, N, Nagata, K, Tanokura, M. | | Deposit date: | 2005-11-21 | | Release date: | 2006-11-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structure of glutamine amidotransferase from Pyrococcus horikoshii OT3
PROC.JPN.ACAD.,SER.B, 81, 2005
|
|
3KKQ
 
 | | Crystal structure of M-Ras P40D in complex with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Ras-related protein M-Ras | | Authors: | Muraoka, S, Shima, F, Liao, J, Ijiri, Y, Matsumoto, K, Ye, M, Inoue, T, Kataoka, T. | | Deposit date: | 2009-11-06 | | Release date: | 2010-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural basis for conformational dynamics of GTP-bound Ras protein
J.Biol.Chem., 285, 2010
|
|
3KKM
 
 | | Crystal structure of H-Ras T35S in complex with GppNHp | | Descriptor: | GTPase HRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Muraoka, S, Shima, F, Liao, J, Ijiri, Y, Matsumoto, K, Ye, M, Inoue, T, Kataoka, T. | | Deposit date: | 2009-11-06 | | Release date: | 2010-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for conformational dynamics of GTP-bound Ras protein
J.Biol.Chem., 285, 2010
|
|
