9GN1
 
 | | Crystal structure of inactive Deacetylase (HdaH) H144A from Klebsiella pneumoniae subsp. ozaenae | | Descriptor: | ACETATE ION, Deacetylase, IMIDAZOLE, ... | | Authors: | Qin, Q, Graf, L.G, Schulze, S, Palm, G.J, Lammers, M. | | Deposit date: | 2024-08-30 | | Release date: | 2024-11-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Distribution and diversity of classical deacylases in bacteria.
Nat Commun, 15, 2024
|
|
9GKX
 
 | | Crystal Structure of Rhizorhabdus wittichii Dimethoate hydrolase (DmhA) in complex with SAHA | | Descriptor: | Dimethoate hydrolase, OCTANEDIOIC ACID HYDROXYAMIDE PHENYLAMIDE, OCTANOIC ACID (CAPRYLIC ACID), ... | | Authors: | Graf, L.G, Lammers, M, Schulze, S, Palm, G.J. | | Deposit date: | 2024-08-26 | | Release date: | 2024-11-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Distribution and diversity of classical deacylases in bacteria.
Nat Commun, 15, 2024
|
|
9GKZ
 
 | | Crystal Structure of Acetylpolyamine amidohydrolase (ApaH) from Pseudomonas sp. M30-35 | | Descriptor: | ACETATE ION, Acetylpolyamine amidohydrolase, CHLORIDE ION, ... | | Authors: | Graf, L.G, Schulze, S, Palm, G.J, Lammers, M. | | Deposit date: | 2024-08-26 | | Release date: | 2024-11-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Distribution and diversity of classical deacylases in bacteria.
Nat Commun, 15, 2024
|
|
9GKV
 
 | | Crystal Structure of Deacetylase (HdaH) from Vibrio cholerae in complex with SAHA | | Descriptor: | ACETATE ION, Histone deacetylase, OCTANEDIOIC ACID HYDROXYAMIDE PHENYLAMIDE, ... | | Authors: | Graf, L.G, Schulze, S, Lammers, M, Palm, G.J. | | Deposit date: | 2024-08-26 | | Release date: | 2024-11-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Distribution and diversity of classical deacylases in bacteria.
Nat Commun, 15, 2024
|
|
9GKW
 
 | | Crystal Structure of Dimethoate hydrolase (DmhA) of Rhizorhabdus wittichii in complex with octanoic acid | | Descriptor: | Dimethoate hydrolase, OCTANOIC ACID (CAPRYLIC ACID), PENTAETHYLENE GLYCOL, ... | | Authors: | Graf, L.G, Schulze, S, Palm, G.J, Lammers, M. | | Deposit date: | 2024-08-26 | | Release date: | 2024-11-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Distribution and diversity of classical deacylases in bacteria.
Nat Commun, 15, 2024
|
|
9GKY
 
 | | Crystal Structure of Histone deacetylase (HdaH) from Vibrio cholerae in complex with decanoic acid | | Descriptor: | DECANOIC ACID, Histone deacetylase, IMIDAZOLE, ... | | Authors: | Graf, L.G, Schulze, S, Palm, G.J, Lammers, M. | | Deposit date: | 2024-08-26 | | Release date: | 2024-11-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Distribution and diversity of classical deacylases in bacteria.
Nat Commun, 15, 2024
|
|
9GL0
 
 | | Crystal Structure of Acetylpolyamine aminohydrolase (ApaH) from Legionella pneumophila | | Descriptor: | Acetylpolyamine aminohydrolase, POTASSIUM ION, ZINC ION | | Authors: | Graf, L.G, Schulze, S, Palm, G.J, Lammers, M. | | Deposit date: | 2024-08-26 | | Release date: | 2024-11-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Distribution and diversity of classical deacylases in bacteria.
Nat Commun, 15, 2024
|
|
9GL1
 
 | | Crystal Structure of Acetylpolyamine aminohydrolase (ApaH) from Legionella cherrii | | Descriptor: | Acetylpolyamine aminohydrolase, POTASSIUM ION, ZINC ION | | Authors: | Graf, L.G, Schulze, S, Palm, G.J, Lammers, M. | | Deposit date: | 2024-08-26 | | Release date: | 2024-11-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Distribution and diversity of classical deacylases in bacteria.
Nat Commun, 15, 2024
|
|
9GLB
 
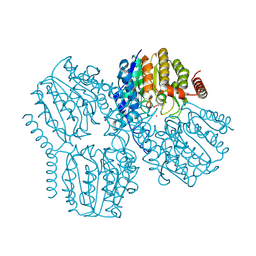 | | Crystal Structure of Deacetylase (HdaH) from Klebsiella pneumoniae subsp. ozaenae | | Descriptor: | ACETATE ION, Deacetylase, GLYCEROL, ... | | Authors: | Qin, C, Graf, L.G, Schulze, S, Palm, G.J, Lammers, M. | | Deposit date: | 2024-08-27 | | Release date: | 2024-11-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Distribution and diversity of classical deacylases in bacteria.
Nat Commun, 15, 2024
|
|
9I4G
 
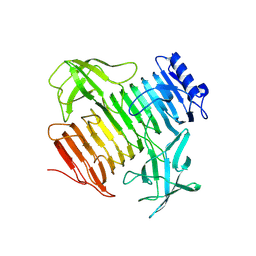 | | Blood Type B-converting alpha-1,3-galactosidase PpaGal from Pedobacter panaciterrae in complex with D-galactose | | Descriptor: | Alpha-1,3-galactosidase B, alpha-D-galactopyranose | | Authors: | Schmoeker, O, Moeller, C, Terholsen, H, Girbardt, B, Palm, G.J, Hoppen, J, Lammers, M, Bornscheuer, U.T. | | Deposit date: | 2025-01-24 | | Release date: | 2025-03-12 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Identification and Protein Engineering of Galactosidases for the Conversion of Blood Type B to Blood Type O.
Chembiochem, 26, 2025
|
|
9I4F
 
 | | Blood Type B-converting alpha-1,3-galactosidase PpaGal from Pedobacter panaciterrae in its apo form | | Descriptor: | 1,2-ETHANEDIOL, Alpha-1,3-galactosidase PpaGal | | Authors: | Schmoeker, O, Moeller, C, Terholsen, H, Girbardt, B, Palm, G.J, Hoppen, J, Lammers, M, Bornscheuer, U.T. | | Deposit date: | 2025-01-24 | | Release date: | 2025-03-12 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Identification and Protein Engineering of Galactosidases for the Conversion of Blood Type B to Blood Type O.
Chembiochem, 26, 2025
|
|
9GKU
 
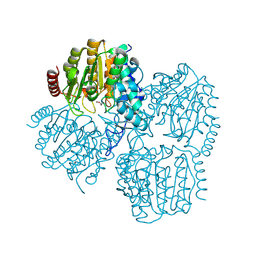 | | Crystal Structure of Propanil hydrolase (PrpH) from Sphingomonas sp. Y57 | | Descriptor: | ACETATE ION, POTASSIUM ION, Propanil hydrolase, ... | | Authors: | Graf, L.G, Lammers, L, Palm, G.J, Schulze, S. | | Deposit date: | 2024-08-26 | | Release date: | 2024-11-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Distribution and diversity of classical deacylases in bacteria.
Nat Commun, 15, 2024
|
|
7CUV
 
 | |
7E30
 
 | | Crystal structure of a novel alpha/beta hydrolase in apo form in complex with citrate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CITRIC ACID, SULFATE ION, ... | | Authors: | Gao, J, Han, X, Zheng, Y.Y, Liu, W.D. | | Deposit date: | 2021-02-07 | | Release date: | 2022-02-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Multiple Substrate Binding Mode-Guided Engineering of a Thermophilic PET Hydrolase.
Acs Catalysis, 12, 2022
|
|
7E31
 
 | | Crystal structure of a novel alpha/beta hydrolase mutant in apo form | | Descriptor: | TRIETHYLENE GLYCOL, alpha/beta hydrolase | | Authors: | Gao, J, Han, X, Zheng, Y.Y, Liu, W.D. | | Deposit date: | 2021-02-07 | | Release date: | 2022-02-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Multiple Substrate Binding Mode-Guided Engineering of a Thermophilic PET Hydrolase.
Acs Catalysis, 12, 2022
|
|
6Z1B
 
 | |
7W1I
 
 | | Crystal structure of carboxylesterase mutant from Thermobifida fusca with C8X and C9C | | Descriptor: | 4-(2-hydroxyethyloxycarbonyl)benzoic acid, Carboxylesterase, bis(2-hydroxyethyl) benzene-1,4-dicarboxylate | | Authors: | Han, X, Gerlis, H, Li, Z, Gao, J, Wei, R, Liu, W. | | Deposit date: | 2021-11-19 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural Insights into (Tere)phthalate-Ester Hydrolysis by a Carboxylesterase and Its Role in Promoting PET Depolymerization
Acs Catalysis, 12, 2022
|
|
7W1J
 
 | | Crystal structure of carboxylesterase from Thermobifida fusca with J1K | | Descriptor: | 4-(2-hydroxyethylcarbamoyl)benzoic acid, Carboxylesterase | | Authors: | Han, X, Gerlis, H, Li, Z, Gao, J, Wei, R, Liu, W. | | Deposit date: | 2021-11-19 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural Insights into (Tere)phthalate-Ester Hydrolysis by a Carboxylesterase and Its Role in Promoting PET Depolymerization
Acs Catalysis, 12, 2022
|
|
7W1K
 
 | | Crystal structure of carboxylesterase from Thermobifida fusca | | Descriptor: | Carboxylesterase | | Authors: | Han, X, Gerlis, H, Li, Z, Gao, J, Wei, R, Liu, W. | | Deposit date: | 2021-11-19 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Structural Insights into (Tere)phthalate-Ester Hydrolysis by a Carboxylesterase and Its Role in Promoting PET Depolymerization
Acs Catalysis, 12, 2022
|
|
7W1L
 
 | | Crystal structure of carboxylesterase mutant from Thermobifida fusca with C8X | | Descriptor: | Carboxylesterase, bis(2-hydroxyethyl) benzene-1,4-dicarboxylate | | Authors: | Han, X, Gerlis, H, Li, Z, Gao, J, Wei, R, Liu, W. | | Deposit date: | 2021-11-19 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structural Insights into (Tere)phthalate-Ester Hydrolysis by a Carboxylesterase and Its Role in Promoting PET Depolymerization
Acs Catalysis, 12, 2022
|
|
7W69
 
 | | Crystal structure of a PSH1 mutant in complex with EDO | | Descriptor: | 1,2-ETHANEDIOL, PSH1 | | Authors: | Gao, J, Lara, P, Li, Z.S, Han, X, Wei, R, Liu, W.D. | | Deposit date: | 2021-12-01 | | Release date: | 2022-09-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Multiple Substrate Binding Mode-Guided Engineering of a Thermophilic PET Hydrolase.
Acs Catalysis, 12, 2022
|
|
7W66
 
 | | Crystal structure of a PSH1 mutant in complex with ligand | | Descriptor: | PSH1, bis(2-hydroxyethyl) benzene-1,4-dicarboxylate | | Authors: | Gao, J, Lara, P, Li, Z.S, Han, X, Wei, R, Liu, W.D. | | Deposit date: | 2021-12-01 | | Release date: | 2022-09-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Multiple Substrate Binding Mode-Guided Engineering of a Thermophilic PET Hydrolase.
Acs Catalysis, 12, 2022
|
|
7W6C
 
 | | Crystal structure of a PSH1 in complex with ligand J1K | | Descriptor: | 4-(2-hydroxyethylcarbamoyl)benzoic acid, PSH1 | | Authors: | Gao, J, Lara, P, Li, Z.S, Han, X, Wei, R, Liu, W.D. | | Deposit date: | 2021-12-01 | | Release date: | 2022-09-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Multiple Substrate Binding Mode-Guided Engineering of a Thermophilic PET Hydrolase.
Acs Catalysis, 12, 2022
|
|
7W6O
 
 | | Crystal structure of a PSH1 in complex with J1K | | Descriptor: | 4-(2-hydroxyethylcarbamoyl)benzoic acid, PSH1 | | Authors: | Gao, J, Lara, P, Li, Z.S, Han, X, Wei, R, Liu, W.D. | | Deposit date: | 2021-12-02 | | Release date: | 2022-09-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Multiple Substrate Binding Mode-Guided Engineering of a Thermophilic PET Hydrolase.
Acs Catalysis, 12, 2022
|
|
7W6Q
 
 | | Crystal structure of a PSH1 in complex with ligand J1K | | Descriptor: | 4-(2-hydroxyethylcarbamoyl)benzoic acid, PSH1 | | Authors: | Gao, J, Lara, P, Li, Z.S, Han, X, Wei, R, Liu, W.D. | | Deposit date: | 2021-12-02 | | Release date: | 2022-09-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Multiple Substrate Binding Mode-Guided Engineering of a Thermophilic PET Hydrolase.
Acs Catalysis, 12, 2022
|
|
