8IEP
 
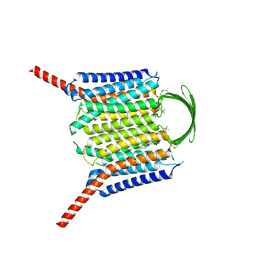 | |
8IEI
 
 | |
6IMP
 
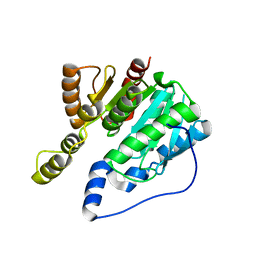 | |
6XJF
 
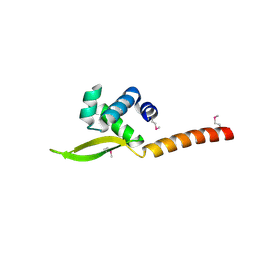 | |
6PLN
 
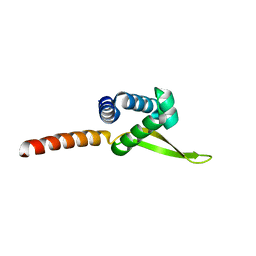 | |
4X0K
 
 | | Engineered Fab fragment specific for EYMPME (EE) peptide | | Descriptor: | Fab fragment heavy chain, Fab fragment light chain | | Authors: | Johnson, J.L, Lieberman, R.L. | | Deposit date: | 2014-11-21 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural and biophysical characterization of an epitope-specific engineered Fab fragment and complexation with membrane proteins: implications for co-crystallization.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
7DGR
 
 | | Activity optimized supercomplex state2 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Jeon, T.J, Lee, S.G, Yoo, S.H, Ryu, J.H, Kim, D.S, Hyun, J.K, Kim, H.M, Ryu, S.E. | | Deposit date: | 2020-11-12 | | Release date: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | A Dynamic Substrate Pool Revealed by cryo-EM of a Lipid-Preserved Respiratory Supercomplex.
Antioxid.Redox Signal., 2022
|
|
7DGQ
 
 | | Activity optimized supercomplex state1 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Jeon, T.J, Lee, S.G, Yoo, S.H, Ryu, J.H, Kim, D.S, Hyun, J.K, Kim, H.M, Ryu, S.E. | | Deposit date: | 2020-11-12 | | Release date: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | A Dynamic Substrate Pool Revealed by cryo-EM of a Lipid-Preserved Respiratory Supercomplex.
Antioxid.Redox Signal., 2022
|
|
7DGZ
 
 | | Activity optimized complex I (closed form) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Jeon, T.J, Lee, S.G, Yoo, S.H, Ryu, J.H, Kim, D.S, Hyun, J.K, Kim, H.M, Ryu, S.E. | | Deposit date: | 2020-11-12 | | Release date: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A Dynamic Substrate Pool Revealed by cryo-EM of a Lipid-Preserved Respiratory Supercomplex.
Antioxid.Redox Signal., 2022
|
|
7DKF
 
 | | Activity optimized supercomplex state4 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Jeon, T.J, Lee, S.G, Yoo, S.H, Ryu, J.H, Kim, D.S, Hyun, J.K, Kim, H.M, Ryu, S.E. | | Deposit date: | 2020-11-24 | | Release date: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (8.3 Å) | | Cite: | A Dynamic Substrate Pool Revealed by cryo-EM of a Lipid-Preserved Respiratory Supercomplex.
Antioxid.Redox Signal., 2022
|
|
7DH0
 
 | | Activity optimized complex I (open form) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Jeon, T.J, Lee, S.G, Yoo, S.H, Ryu, J.H, Kim, D.S, Hyun, J.K, Kim, H.M, Ryu, S.E. | | Deposit date: | 2020-11-12 | | Release date: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | A Dynamic Substrate Pool Revealed by cryo-EM of a Lipid-Preserved Respiratory Supercomplex.
Antioxid.Redox Signal., 2022
|
|
7DGS
 
 | | Activity optimized supercomplex state3 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Jeon, T.J, Lee, S.G, Yoo, S.H, Ryu, J.H, Kim, D.S, Hyun, J.K, Kim, H.M, Ryu, S.E. | | Deposit date: | 2020-11-12 | | Release date: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | A Dynamic Substrate Pool Revealed by cryo-EM of a Lipid-Preserved Respiratory Supercomplex.
Antioxid.Redox Signal., 2022
|
|
8IEQ
 
 | | Cryo-EM structure of G-protein free GPR156 | | Descriptor: | Probable G-protein coupled receptor 156, [(2R)-3-[(E)-hexadec-9-enoyl]oxy-2-octadecanoyloxy-propyl] 2-(trimethylazaniumyl)ethyl phosphate | | Authors: | Shin, J, Park, J, Cho, Y. | | Deposit date: | 2023-02-15 | | Release date: | 2024-02-14 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Constitutive activation mechanism of a class C GPCR.
Nat.Struct.Mol.Biol., 31, 2024
|
|
6RAO
 
 | |
6RGL
 
 | |
6RAP
 
 | |
6RC8
 
 | |
6RBK
 
 | |
6RBN
 
 | |
5YPC
 
 | | p62/SQSTM1 ZZ domain with Phe-peptide | | Descriptor: | 78 kDa glucose-regulated protein,Sequestosome-1, ZINC ION | | Authors: | Kwon, D.H, Kim, L, Song, H.K. | | Deposit date: | 2017-11-01 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.962 Å) | | Cite: | Insights into degradation mechanism of N-end rule substrates by p62/SQSTM1 autophagy adapter.
Nat Commun, 9, 2018
|
|
5YPB
 
 | | p62/SQSTM1 ZZ domain with His-peptide | | Descriptor: | 78 kDa glucose-regulated protein,Sequestosome-1, ZINC ION | | Authors: | Kwon, D.H, Kim, L, Song, H.K. | | Deposit date: | 2017-11-01 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Insights into degradation mechanism of N-end rule substrates by p62/SQSTM1 autophagy adapter.
Nat Commun, 9, 2018
|
|
5YPH
 
 | | p62/SQSTM1 ZZ domain with Ile-peptide | | Descriptor: | 78 kDa glucose-regulated protein,Sequestosome-1, ZINC ION | | Authors: | Kwon, D.H, Kim, L, Song, H.K. | | Deposit date: | 2017-11-01 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.629 Å) | | Cite: | Insights into degradation mechanism of N-end rule substrates by p62/SQSTM1 autophagy adapter.
Nat Commun, 9, 2018
|
|
5YP8
 
 | | p62/SQSTM1 ZZ domain with Arg-peptide | | Descriptor: | 78 kDa glucose-regulated protein,Sequestosome-1, ZINC ION | | Authors: | Kwon, D.H, Kim, L, Song, H.K. | | Deposit date: | 2017-11-01 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.448 Å) | | Cite: | Insights into degradation mechanism of N-end rule substrates by p62/SQSTM1 autophagy adapter.
Nat Commun, 9, 2018
|
|
5YPG
 
 | | p62/SQSTM1 ZZ domain with Leu-peptide | | Descriptor: | 78 kDa glucose-regulated protein,Sequestosome-1, ZINC ION | | Authors: | Kwon, D.H, Kim, L, Song, H.K. | | Deposit date: | 2017-11-01 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | Insights into degradation mechanism of N-end rule substrates by p62/SQSTM1 autophagy adapter.
Nat Commun, 9, 2018
|
|
5YPF
 
 | | p62/SQSTM1 ZZ domain with Trp-peptide | | Descriptor: | 78 kDa glucose-regulated protein,Sequestosome-1, ZINC ION | | Authors: | Kwon, D.H, Kim, L, Song, H.K. | | Deposit date: | 2017-11-01 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.951 Å) | | Cite: | Insights into degradation mechanism of N-end rule substrates by p62/SQSTM1 autophagy adapter.
Nat Commun, 9, 2018
|
|
