4WJM
 
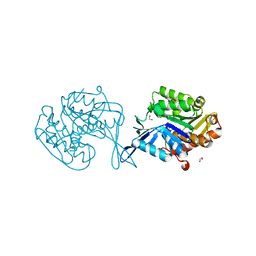 | |
5VVW
 
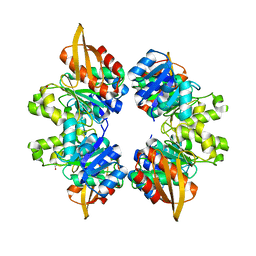 | |
5UUI
 
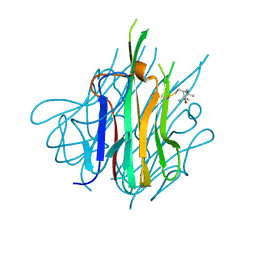 | | Crystal Structure of Spin-Labeled T77C TNFa | | Descriptor: | S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate, Tumor necrosis factor | | Authors: | Horanyi, P.S, Dranow, D.M, Ceska, T. | | Deposit date: | 2017-02-16 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Natural Conformational Sampling of Human TNF alpha Visualized by Double Electron-Electron Resonance.
Biophys. J., 113, 2017
|
|
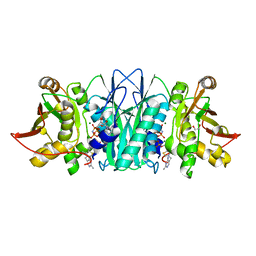
5DD7
 
 | |
8CSO
 
 | |
6P81
 
 | |
6PBL
 
 | |
6PTR
 
 | |
6Q1Y
 
 | |
1RYQ
 
 | | Putative DNA-directed RNA polymerase, subunit e'' from Pyrococcus Furiosus Pfu-263306-001 | | Descriptor: | DNA-directed RNA polymerase, subunit e'', ZINC ION | | Authors: | Liu, Z.-J, Chen, L, Tempel, W, Shah, A, Arendall III, W.B, Rose, J.P, Brereton, P.S, Izumi, M, Jenney Jr, F.E, Lee, H.S, Poole II, F.L, Shah, C, Sugar, F.J, Adams, M.W.W, Richardson, D.C, Richardson, J.S, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2003-12-22 | | Release date: | 2004-08-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Parameter-space screening: a powerful tool for high-throughput crystal structure determination.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
6PTG
 
 | |
6Q10
 
 | |
6PTV
 
 | |
6PTH
 
 | |
6PZJ
 
 | |
4U83
 
 | |
4XGI
 
 | |
5VPR
 
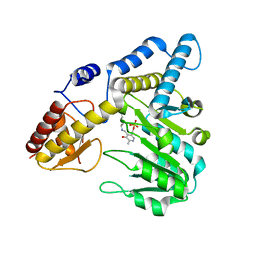 | |
5VMT
 
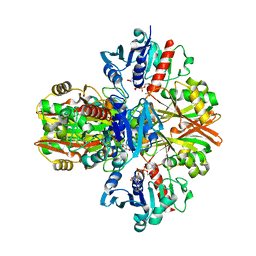 | |
5CC8
 
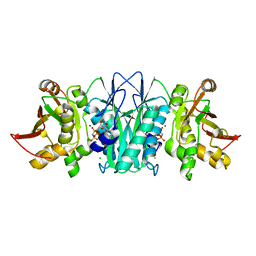 | |
5CM7
 
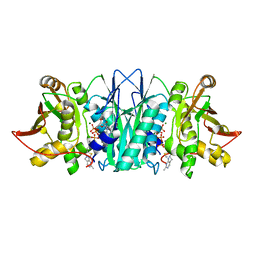 | |
6CFP
 
 | |
6CKT
 
 | | Crystal structure of 2,3,4,5-tetrahydropyridine-2,6-dicarboxylate N-succinyltransferase from Legionella pneumophila Philadelphia 1 | | Descriptor: | 1,2-ETHANEDIOL, 2,3,4,5-tetrahydropyridine-2,6-dicarboxylate N-succinyltransferase | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2018-02-28 | | Release date: | 2018-03-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of 2,3,4,5-tetrahydropyridine-2,6-dicarboxylate N-succinyltransferase from Legionella pneumophila Philadelphia 1
to be published
|
|
6CU3
 
 | |
6CK0
 
 | |
