3W26
 
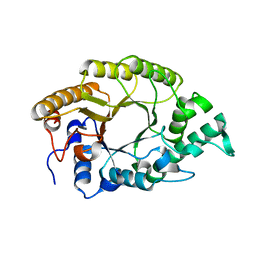 | | The high-resolution crystal structure of TsXylA, intracellular xylanase from /Thermoanaerobacterium saccharolyticum JW/SL-YS485/: the complex of the E146A mutant with xylotriose | | Descriptor: | Glycoside hydrolase family 10, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Han, X, Gao, J, Shang, N, Huang, C.-H, Ko, T.-P, Zhu, Z, Wiegel, J, Shao, W, Guo, R.-T. | | Deposit date: | 2012-11-27 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and functional analyses of catalytic domain of GH10 xylanase from Thermoanaerobacterium saccharolyticum JW/SL-YS485
Proteins, 81, 2013
|
|
3W24
 
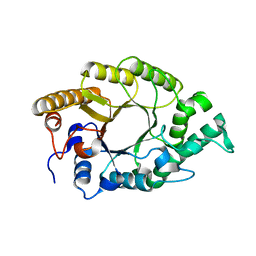 | | The high-resolution crystal structure of TsXylA, intracellular xylanase from Thermoanaerobacterium saccharolyticum JW/SL-YS485 | | Descriptor: | Glycoside hydrolase family 10 | | Authors: | Han, X, Gao, J, Shang, N, Huang, C.-H, Ko, T.-P, Zhu, Z, Wiegel, J, Shao, W, Guo, R.-T. | | Deposit date: | 2012-11-27 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural and functional analyses of catalytic domain of GH10 xylanase from Thermoanaerobacterium saccharolyticum JW/SL-YS485
Proteins, 81, 2013
|
|
8I0T
 
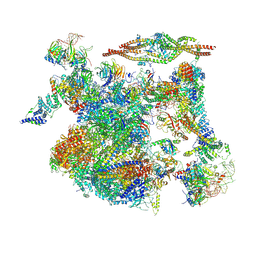 | | The cryo-EM structure of human Bact-III complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, Cell division cycle 5-like protein, Crooked neck-like protein 1, ... | | Authors: | Zhan, X, Lu, Y, Shi, Y. | | Deposit date: | 2023-01-11 | | Release date: | 2024-07-31 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular basis for the activation of human spliceosome.
Nat Commun, 15, 2024
|
|
8I0P
 
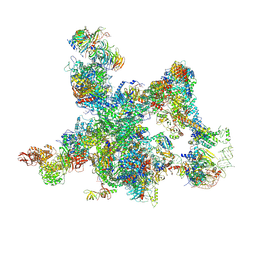 | | The cryo-EM structure of human pre-Bact complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, BUD13 homolog, Cell division cycle 5-like protein, ... | | Authors: | Zhan, X, Lu, Y, Shi, Y. | | Deposit date: | 2023-01-11 | | Release date: | 2024-07-31 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular basis for the activation of human spliceosome.
Nat Commun, 15, 2024
|
|
8I0R
 
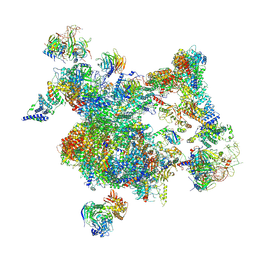 | | The cryo-EM structure of human Bact-I complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, BUD13 homolog, Cell division cycle 5-like protein, ... | | Authors: | Zhan, X, Lu, Y, Shi, Y. | | Deposit date: | 2023-01-11 | | Release date: | 2024-07-31 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular basis for the activation of human spliceosome.
Nat Commun, 15, 2024
|
|
8I0V
 
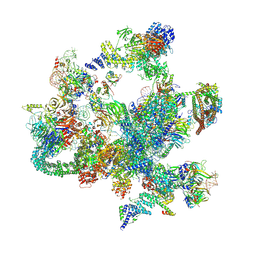 | | The cryo-EM structure of human post-Bact complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, Cell division cycle 5-like protein, Crooked neck-like protein 1, ... | | Authors: | Zhan, X, Lu, Y, Shi, Y. | | Deposit date: | 2023-01-11 | | Release date: | 2024-07-31 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular basis for the activation of human spliceosome.
Nat Commun, 15, 2024
|
|
8I0S
 
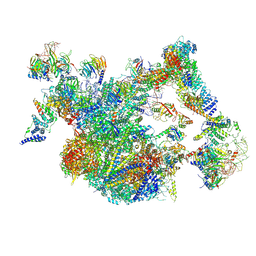 | | The cryo-EM structure of human Bact-II complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, Cell division cycle 5-like protein, Crooked neck-like protein 1, ... | | Authors: | Zhan, X, Lu, Y, Shi, Y. | | Deposit date: | 2023-01-11 | | Release date: | 2024-07-31 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Molecular basis for the activation of human spliceosome.
Nat Commun, 15, 2024
|
|
8I0W
 
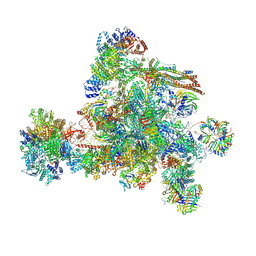 | | The cryo-EM structure of human C complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, Cell division cycle 5-like protein, Coiled-coil domain-containing protein 94, ... | | Authors: | Zhan, X, Lu, Y, Shi, Y. | | Deposit date: | 2023-01-11 | | Release date: | 2024-07-31 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular basis for the activation of human spliceosome.
Nat Commun, 15, 2024
|
|
8I0U
 
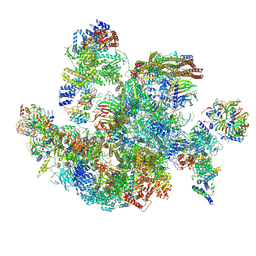 | | The cryo-EM structure of human Bact-IV complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, Cell division cycle 5-like protein, Crooked neck-like protein 1, ... | | Authors: | Zhan, X, Lu, Y, Shi, Y. | | Deposit date: | 2023-01-11 | | Release date: | 2024-07-31 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular basis for the activation of human spliceosome.
Nat Commun, 15, 2024
|
|
7F8Y
 
 | | Crystal structure of the cholecystokinin receptor CCKAR in complex with devazepide | | Descriptor: | N-[(3S)-1-methyl-2-oxidanylidene-5-phenyl-3H-1,4-benzodiazepin-3-yl]-1H-indole-2-carboxamide, fusion protein of Cholecystokinin receptor type A and Endolysin | | Authors: | Zhang, X, He, C, Wang, M, Zhou, Q, Yang, D, Zhu, Y, Wu, B, Zhao, Q. | | Deposit date: | 2021-07-02 | | Release date: | 2021-10-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of the human cholecystokinin receptors bound to agonists and antagonists.
Nat.Chem.Biol., 17, 2021
|
|
7F8U
 
 | | Crystal structure of the cholecystokinin receptor CCKAR in complex with lintitript | | Descriptor: | 2-[2-[[4-(2-chlorophenyl)-1,3-thiazol-2-yl]carbamoyl]indol-1-yl]ethanoic acid, Fusion protein of Cholecystokinin receptor type A and Endolysin | | Authors: | Zhang, X, He, C, Wang, M, Zhou, Q, Yang, D, Zhu, Y, Wu, B, Zhao, Q. | | Deposit date: | 2021-07-02 | | Release date: | 2021-10-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of the human cholecystokinin receptors bound to agonists and antagonists.
Nat.Chem.Biol., 17, 2021
|
|
7F8V
 
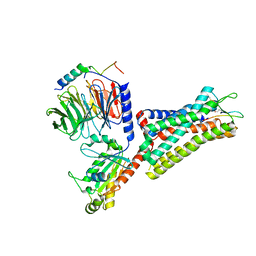 | | Cryo-EM structure of the cholecystokinin receptor CCKBR in complex with gastrin-17 and Gi | | Descriptor: | Gastrin-17, Gastrin/cholecystokinin type B receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhang, X, He, C, Wang, M, Zhou, Q, Yang, D, Zhu, Y, Wu, B, Zhao, Q. | | Deposit date: | 2021-07-02 | | Release date: | 2021-10-13 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures of the human cholecystokinin receptors bound to agonists and antagonists.
Nat.Chem.Biol., 17, 2021
|
|
7F8W
 
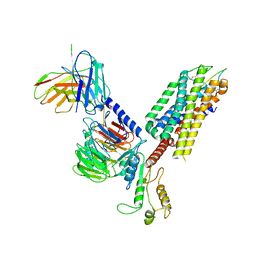 | | Cryo-EM structure of the cholecystokinin receptor CCKBR in complex with gastrin-17 and Gq | | Descriptor: | Gastrin-17, Gastrin/cholecystokinin type B receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhang, X, He, C, Wang, M, Zhou, Q, Yang, D, Zhu, Y, Wu, B, Zhao, Q. | | Deposit date: | 2021-07-02 | | Release date: | 2021-10-13 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structures of the human cholecystokinin receptors bound to agonists and antagonists.
Nat.Chem.Biol., 17, 2021
|
|
7F8X
 
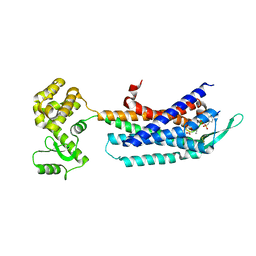 | | Crystal structure of the cholecystokinin receptor CCKAR in complex with NN9056 | | Descriptor: | ASP-SMF-NLE-GLY-TRP-NLE-OEM-MEA-NH2 (NN9056), Cholecystokinin receptor type A,Endolysin | | Authors: | Zhang, X, He, C, Wang, M, Zhou, Q, Yang, D, Zhu, Y, Wu, B, Zhao, Q. | | Deposit date: | 2021-07-02 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of the human cholecystokinin receptors bound to agonists and antagonists.
Nat.Chem.Biol., 17, 2021
|
|
1A83
 
 | | INTRAMOLECULAR I-MOTIF, NMR, 6 STRUCTURES | | Descriptor: | DNA (5'-(MCY)CTTTCCTTTACCTTTCC-3') | | Authors: | Han, X, Leroy, J.L, Gueron, M. | | Deposit date: | 1998-04-01 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | An intramolecular i-motif: the solution structure and base-pair opening kinetics of d(5mCCT3CCT3ACCT3CC).
J.Mol.Biol., 278, 1998
|
|
7X1Z
 
 | | Structure of the phosphorylation-site double mutant S431E/T432E of the KaiC circadian clock protein | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Circadian clock oscillator protein KaiC, MAGNESIUM ION | | Authors: | Han, X, Zhang, D.L, Hong, L, Yu, D.Q, Wu, Z.L, Yang, T, Rust, M.J, Tu, Y.H, Ouyang, Q. | | Deposit date: | 2022-02-25 | | Release date: | 2023-04-19 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Determining subunit-subunit interaction from statistics of cryo-EM images: observation of nearest-neighbor coupling in a circadian clock protein complex
Nat Commun, 14, 2023
|
|
7X1Y
 
 | | Structure of the phosphorylation-site double mutant S431A/T432A of the KaiC circadian clock protein | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Circadian clock oscillator protein KaiC, MAGNESIUM ION | | Authors: | Han, X, Zhang, D.L, Hong, L, Yu, D.Q, Wu, Z.L, Yang, T, Rust, M.J, Tu, Y.H, Ouyang, Q. | | Deposit date: | 2022-02-25 | | Release date: | 2023-04-26 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Determining subunit-subunit interaction from statistics of cryo-EM images: observation of nearest-neighbor coupling in a circadian clock protein complex
Nat Commun, 14, 2023
|
|
1IBN
 
 | |
1IBO
 
 | |
8IJ8
 
 | |
8IJ6
 
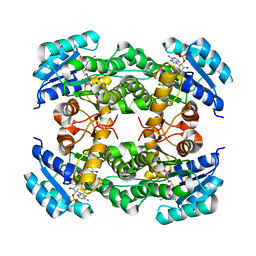 | | Crystal structure of alcohol dehydrogenase from Burkholderia gladioli with NADP | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative short-chain dehydrogenases/reductase family protein | | Authors: | Han, X, Mei, Z.L, Liu, W.D, Sun, Z.T, Ma, J.A. | | Deposit date: | 2023-02-26 | | Release date: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structure of alcohol dehydrogenase from Burkholderia gladioli with NADP
To Be Published
|
|
8IJ7
 
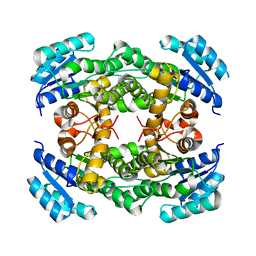 | |
8IJG
 
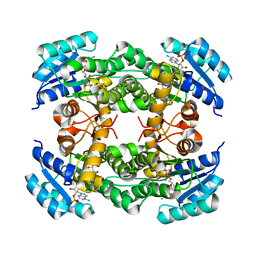 | | Crystal structure of alcohol dehydrogenase M5 from Burkholderia gladioli with NADP | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative short-chain dehydrogenases/reductase family protein | | Authors: | Han, X, Mei, Z.L, Liu, W.D, Sun, Z.T, Ma, J.A. | | Deposit date: | 2023-02-27 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal structure of alcohol dehydrogenase from Burkholderia gladioli with NADP
To Be Published
|
|
7YKP
 
 | | Crystal structure of a novel alpha/beta hydrolase from thermomonospora curvata with glycerol | | Descriptor: | GLYCEROL, Triacylglycerol lipase | | Authors: | Han, X, Jian, G, Bornscheuer, U.T, Wei, R, Liu, W. | | Deposit date: | 2022-07-23 | | Release date: | 2023-07-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Crystal structure of a novel alpha/beta hydrolase from thermomonospora curvata with glycerol
To Be Published
|
|
7YKO
 
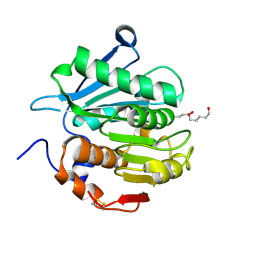 | | Crystal structure of a novel alpha/beta hydrolase mutant from thermomonospora curvata in complex with pentane-1,5-diol | | Descriptor: | Triacylglycerol lipase, pentane-1,5-diol | | Authors: | Han, X, Jian, G, Bornscheuer, U.T, Wei, R, Liu, W. | | Deposit date: | 2022-07-23 | | Release date: | 2023-07-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystal structure of a novel alpha/beta hydrolase mutant from thermomonospora curvata in complex with pentane-1,5-diol
To Be Published
|
|
