8HIC
 
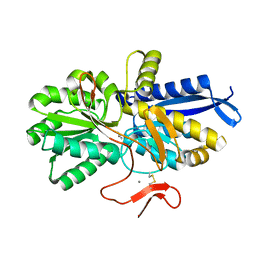 | | Crystal structure of UrtA from Prochlorococcus marinus str. MIT 9313 in complex with urea and calcium | | Descriptor: | CALCIUM ION, Putative urea ABC transporter, substrate binding protein, ... | | Authors: | Zhang, Y.Z, Wang, P, Wang, C. | | Deposit date: | 2022-11-19 | | Release date: | 2023-11-22 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and molecular basis for urea recognition by Prochlorococcus.
J.Biol.Chem., 299, 2023
|
|
6J76
 
 | | Structure of 3,6-anhydro-L-galactose Dehydrogenase in Complex with NAP | | Descriptor: | Aldehyde dehydrogenase A, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Li, P.Y, Wang, Y, Chen, X.L, Zhang, Y.Z. | | Deposit date: | 2019-01-17 | | Release date: | 2020-01-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.368 Å) | | Cite: | 3,6-Anhydro-L-Galactose Dehydrogenase VvAHGD is a Member of a New Aldehyde Dehydrogenase Family and Catalyzes by a Novel Mechanism with Conformational Switch of Two Catalytic Residues Cysteine 282 and Glutamate 248.
J.Mol.Biol., 432, 2020
|
|
6J75
 
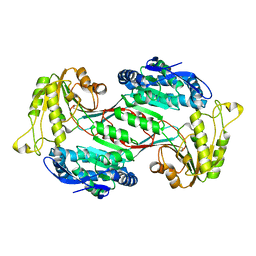 | | Structure of 3,6-anhydro-L-galactose Dehydrogenase | | Descriptor: | Aldehyde dehydrogenase A | | Authors: | Li, P.Y, Wang, Y, Chen, X.L, Zhang, Y.Z. | | Deposit date: | 2019-01-17 | | Release date: | 2020-01-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.695 Å) | | Cite: | 3,6-Anhydro-L-Galactose Dehydrogenase VvAHGD is a Member of a New Aldehyde Dehydrogenase Family and Catalyzes by a Novel Mechanism with Conformational Switch of Two Catalytic Residues Cysteine 282 and Glutamate 248.
J.Mol.Biol., 432, 2020
|
|
7DDY
 
 | |
7CZH
 
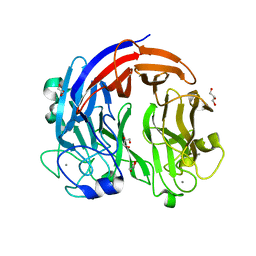 | | PL24 ulvan lyase-Uly1 | | Descriptor: | CALCIUM ION, GLYCEROL, Uly1 | | Authors: | Zhang, Y.Z, Chen, X.L, Dong, F, Xu, F. | | Deposit date: | 2020-09-08 | | Release date: | 2021-04-07 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.104 Å) | | Cite: | Mechanistic Insights into Substrate Recognition and Catalysis of a New Ulvan Lyase of Polysaccharide Lyase Family 24.
Appl.Environ.Microbiol., 87, 2021
|
|
