4Q1K
 
 | |
4Q1J
 
 | | Structure and mechanism of a dehydratase/decarboxylase enzyme couple involved in polyketide beta-branching | | Descriptor: | 1,2-ETHANEDIOL, Polyketide biosynthesis enoyl-CoA isomerase PksI, SODIUM ION | | Authors: | Nair, A.V, Race, P.R, Till, M. | | Deposit date: | 2014-04-03 | | Release date: | 2015-05-06 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structure and mechanism of a dehydratase/decarboxylase enzyme couple involved in polyketide beta-methyl branch incorporation.
Sci Rep, 10, 2020
|
|
4Q1G
 
 | | Structure and mechanism of a dehydratase/decarboxylase enzyme couple involved in polyketide beta-branching | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Polyketide biosynthesis enoyl-CoA isomerase PksI | | Authors: | Nair, A.V, Race, P.R, Till, M. | | Deposit date: | 2014-04-03 | | Release date: | 2015-05-06 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and mechanism of a dehydratase/decarboxylase enzyme couple involved in polyketide beta-methyl branch incorporation.
Sci Rep, 10, 2020
|
|
4Q1I
 
 | |
8CBW
 
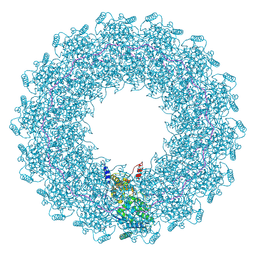 | | CryoEM structure of the Hendra henipavirus nucleocapsid sauronoid assembly monomer | | Descriptor: | Nucleocapsid, RNA (5'-R(P*UP*UP*UP*UP*UP*U)-3') | | Authors: | Passchier, T.C, Maskell, D.P, Edwards, T.A, Barr, J.N. | | Deposit date: | 2023-01-26 | | Release date: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.485 Å) | | Cite: | The cryoEM structure of the Hendra henipavirus nucleoprotein reveals insights into paramyxoviral nucleocapsid architectures.
Sci Rep, 14, 2024
|
|
8C4H
 
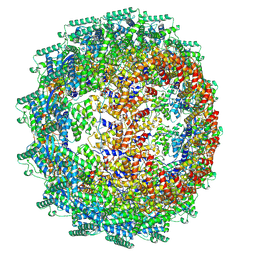 | | CryoEM structure of the Hendra henipavirus nucleocapsid sauronoid assembly multimer | | Descriptor: | Nucleocapsid, RNA (84-MER) | | Authors: | Passchier, T.C, Maskell, D.P, Edwards, T.A, Barr, J.N. | | Deposit date: | 2023-01-04 | | Release date: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.485 Å) | | Cite: | The cryoEM structure of the Hendra henipavirus nucleoprotein reveals insights into paramyxoviral nucleocapsid architectures.
Sci Rep, 14, 2024
|
|
