5OK0
 
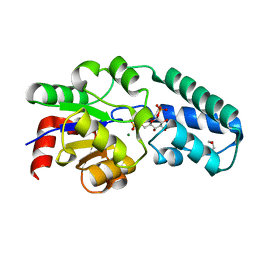 | | Structure of the D10N mutant of beta-phosphoglucomutase from Lactococcus lactis trapped with native reaction intermediate beta-glucose 1,6-bisphosphate to 2.2A resolution. | | Descriptor: | 1,3-PROPANDIOL, 1,6-di-O-phosphono-beta-D-glucopyranose, Beta-phosphoglucomutase, ... | | Authors: | Robertson, A.J, Bisson, C. | | Deposit date: | 2017-07-25 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | van der Waals Contact between Nucleophile and Transferring Phosphorus Is Insufficient To Achieve Enzyme Transition-State Architecture
Acs Catalysis, 2018
|
|
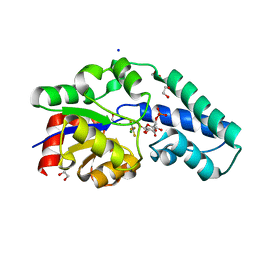
5OK2
 
 | |
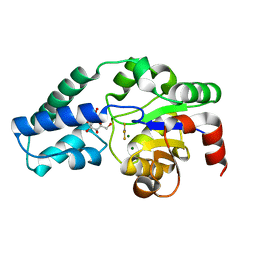
5O6R
 
 | |
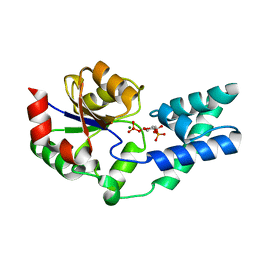
5O6P
 
 | |
5OJZ
 
 | |
6GIA
 
 | | Crystal structure of pentaerythritol tetranitrate reductase (PETNR) mutant I107A | | Descriptor: | ACETATE ION, FLAVIN MONONUCLEOTIDE, Pentaerythritol tetranitrate reductase | | Authors: | Levy, C.W. | | Deposit date: | 2018-05-10 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Nonequivalence of Second Sphere "Noncatalytic" Residues in Pentaerythritol Tetranitrate Reductase in Relation to Local Dynamics Linked to H-Transfer in Reactions with NADH and NADPH Coenzymes.
Acs Catalysis, 8, 2018
|
|
6GI8
 
 | | Crystal structure of pentaerythritol tetranitrate reductase (PETNR) mutant L25A | | Descriptor: | ACETATE ION, FLAVIN MONONUCLEOTIDE, Pentaerythritol tetranitrate reductase | | Authors: | Levy, C.W. | | Deposit date: | 2018-05-10 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Nonequivalence of Second Sphere "Noncatalytic" Residues in Pentaerythritol Tetranitrate Reductase in Relation to Local Dynamics Linked to H-Transfer in Reactions with NADH and NADPH Coenzymes.
Acs Catalysis, 8, 2018
|
|
6GI7
 
 | | Crystal structure of pentaerythritol tetranitrate reductase (PETNR) mutant L25I | | Descriptor: | ACETATE ION, FLAVIN MONONUCLEOTIDE, Pentaerythritol tetranitrate reductase | | Authors: | Levy, C.W. | | Deposit date: | 2018-05-10 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Nonequivalence of Second Sphere "Noncatalytic" Residues in Pentaerythritol Tetranitrate Reductase in Relation to Local Dynamics Linked to H-Transfer in Reactions with NADH and NADPH Coenzymes.
Acs Catalysis, 8, 2018
|
|
5OK1
 
 | |
6GI9
 
 | | Crystal structure of pentaerythritol tetranitrate reductase (PETNR) mutant I107L | | Descriptor: | ACETATE ION, FLAVIN MONONUCLEOTIDE, Pentaerythritol tetranitrate reductase | | Authors: | Levy, C.W. | | Deposit date: | 2018-05-10 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Nonequivalence of Second Sphere "Noncatalytic" Residues in Pentaerythritol Tetranitrate Reductase in Relation to Local Dynamics Linked to H-Transfer in Reactions with NADH and NADPH Coenzymes.
Acs Catalysis, 8, 2018
|
|
2RSU
 
 | | Alternative structure of Ubiquitin | | Descriptor: | Ubiquitin | | Authors: | Kitazawa, S, Kameda, T, Yagi-Utsumi, M, Kato, K, Kitahara, R. | | Deposit date: | 2012-06-15 | | Release date: | 2013-03-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Q41N Variant of Ubiquitin as a Model for the Alternatively Folded N2 State of Ubiquitin
Biochemistry, 52, 2013
|
|
4C4S
 
 | |
4C4R
 
 | |
4C4T
 
 | |
2XE6
 
 | | The complete reaction cycle of human phosphoglycerate kinase: The open binary complex with 3PG | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, PHOSPHOGLYCERATE KINASE 1 | | Authors: | Cliff, M.J, Baxter, N.J, Blackburn, G.M, Merli, A, Vas, M, Waltho, J.P, Bowler, M.W. | | Deposit date: | 2010-05-11 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | A Spring Loaded Release Mechanism Regulates Domain Movement and Catalysis in Phosphoglycerate Kinase.
J.Biol.Chem., 286, 2011
|
|
2XE8
 
 | | The complete reaction cycle of human phosphoglycerate kinase: The open ternary complex with 3PG and AMP-PNP | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, PHOSPHOGLYCERATE KINASE 1, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Cliff, M.J, Baxter, N.J, Blackburn, G.M, Merli, A, Vas, M, Waltho, J.P, Bowler, M.W. | | Deposit date: | 2010-05-11 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | A Spring Loaded Release Mechanism Regulates Domain Movement and Catalysis in Phosphoglycerate Kinase.
J.Biol.Chem., 286, 2011
|
|
2XE7
 
 | | The complete reaction cycle of human phosphoglycerate kinase: The open ternary complex with 3PG and ADP | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, ADENOSINE-5'-DIPHOSPHATE, PHOSPHOGLYCERATE KINASE 1 | | Authors: | Cliff, M.J, Baxter, N.J, Blackburn, G.M, Merli, A, Vas, M, Waltho, J.P, Bowler, M.W. | | Deposit date: | 2010-05-11 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Spring Loaded Release Mechanism Regulates Domain Movement and Catalysis in Phosphoglycerate Kinase.
J.Biol.Chem., 286, 2011
|
|
