5B5H
 
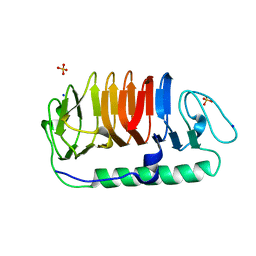 | | Hydrophobic ice-binding site confer hyperactivity on antifreeze protein from a snow mold fungus | | Descriptor: | Antifreeze protein, SODIUM ION, SULFATE ION | | Authors: | Cheng, J, Hanada, Y, Miura, A, Tsuda, S, Kondo, H. | | Deposit date: | 2016-05-06 | | Release date: | 2016-09-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Hydrophobic ice-binding sites confer hyperactivity of an antifreeze protein from a snow mold fungus.
Biochem.J., 473, 2016
|
|
7DC5
 
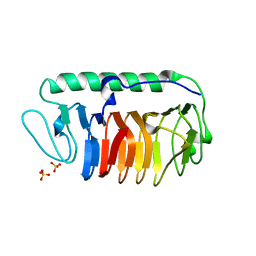 | | Crystal structure of fungal antifreeze protein with intermediate activity | | Descriptor: | Antifreeze protein, SULFATE ION | | Authors: | Khan, N.M.M.U, Arai, T, Tsuda, S, Kondo, H. | | Deposit date: | 2020-10-23 | | Release date: | 2021-10-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Characterization of microbial antifreeze protein with intermediate activity suggests that a bound-water network is essential for hyperactivity.
Sci Rep, 11, 2021
|
|
7DDB
 
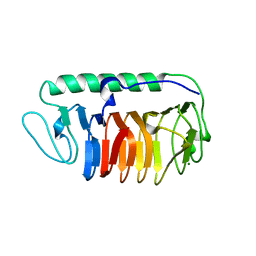 | | Crystal structure of fungal antifreeze protein with intermediate activity | | Descriptor: | Antifreeze protein, MAGNESIUM ION | | Authors: | Khan, N.M.M.U, Arai, T, Tsuda, S, Kondo, H. | | Deposit date: | 2020-10-28 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Characterization of microbial antifreeze protein with intermediate activity suggests that a bound-water network is essential for hyperactivity.
Sci Rep, 11, 2021
|
|
2ZIB
 
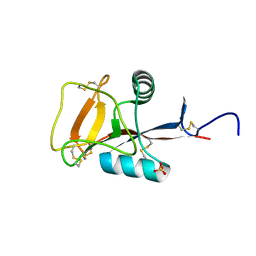 | | Crystal structure analysis of calcium-independent type II antifreeze protein | | Descriptor: | SULFATE ION, Type II antifreeze protein | | Authors: | Nishimiya, Y, Sato, R, Kondo, H, Noro, N, Sugimoto, H, Suzuki, M, Tsuda, S. | | Deposit date: | 2008-02-14 | | Release date: | 2008-08-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Crystal structure and mutational analysis of Ca2+-independent type II antifreeze protein from longsnout poacher, Brachyopsis rostratus
J.Mol.Biol., 382, 2008
|
|
3A0F
 
 | | The crystal structure of Geotrichum sp. M128 xyloglucanase | | Descriptor: | Xyloglucanase | | Authors: | Yaoi, K, Kondo, H, Hiyoshi, A, Noro, N, Sugimoto, H, Tsuda, S, Miyazaki, K. | | Deposit date: | 2009-03-16 | | Release date: | 2009-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of a xyloglucan-specific endo-beta-1,4-glucanase from Geotrichum sp. M128 xyloglucanase reveals a key amino acid residue for substrate specificity
Febs J., 276, 2009
|
|
2D56
 
 | | Solution Structure of ASABF, Antibacterial Peptide Isolated from a Nematode, Ascaris Suum | | Descriptor: | ASABF | | Authors: | Nakano, M, Aizawa, T, Kamiya, M, Miura, K, Kumaki, Y, Demura, M, Tsuda, S, Kawano, K. | | Deposit date: | 2005-10-28 | | Release date: | 2006-11-14 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure and function of ASABFd18c, antibacterial peptide isolated from a nematode, asucaris suum
To be Published
|
|
2DCZ
 
 | | Thermal Stabilization of Bacillus subtilis Family-11 Xylanase By Directed Evolution | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Endo-1,4-beta-xylanase A, SULFATE ION | | Authors: | Kondo, H, Miyazaki, K, Takenouchi, M, Noro, N, Suzuki, M, Tsuda, S. | | Deposit date: | 2006-01-18 | | Release date: | 2006-02-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Thermal Stabilization of Bacillus subtilis Family-11 Xylanase by Directed Evolution
J.Biol.Chem., 281, 2006
|
|
2DCY
 
 | | Crystal structure of Bacillus subtilis family-11 xylanase | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, D(-)-TARTARIC ACID, Endo-1,4-beta-xylanase A, ... | | Authors: | Kondo, H, Miyazaki, K, Takenouchi, M, Noro, N, Suzuki, M, Tsuda, S. | | Deposit date: | 2006-01-18 | | Release date: | 2006-02-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Thermal Stabilization of Bacillus subtilis Family-11 Xylanase by Directed Evolution
J.Biol.Chem., 281, 2006
|
|
2CZQ
 
 | | A novel cutinase-like protein from Cryptococcus sp. | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, cutinase-like protein | | Authors: | Masaki, K, Kamini, N.R, Ikeda, H, Iefuji, H, Kondo, H, Suzuki, M, Tsuda, S. | | Deposit date: | 2005-07-14 | | Release date: | 2006-07-14 | | Last modified: | 2012-06-13 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Crystal structure and enhanced activity of a cutinase-like enzyme from Cryptococcus sp. strain S-2
Proteins, 77, 2009
|
|
1TNQ
 
 | |
1TNP
 
 | |
3NLA
 
 | | NMR STRUCTURE OF THE N-TERMINAL DOMAIN WITH A LINKER PORTION OF ANTARCTIC EEL POUT ANTIFREEZE PROTEIN RD3, 40 STRUCTURES | | Descriptor: | ANTIFREEZE PROTEIN RD3 TYPE III | | Authors: | Miura, K, Ohgiya, S, Hoshino, T, Nemoto, N, Hikichi, K, Tsuda, S. | | Deposit date: | 1998-02-24 | | Release date: | 1999-02-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the binding of a globular antifreeze protein to ice.
Nature, 384, 1996
|
|
3RDN
 
 | | NMR STRUCTURE OF THE N-TERMINAL DOMAIN WITH A LINKER PORTION OF ANTARCTIC EEL POUT ANTIFREEZE PROTEIN RD3, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | ANTIFREEZE PROTEIN RD3 TYPE III | | Authors: | Miura, K, Ohgiya, S, Hoshino, T, Nemoto, N, Hikichi, K, Tsuda, S. | | Deposit date: | 1998-02-24 | | Release date: | 1999-02-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the binding of a globular antifreeze protein to ice.
Nature, 384, 1996
|
|
2EBS
 
 | | Crystal Structure Anaalysis of Oligoxyloglucan reducing-end-specific cellobiohydrolase (OXG-RCBH) D465N Mutant Complexed with a Xyloglucan Heptasaccharide | | Descriptor: | Oligoxyloglucan reducing end-specific cellobiohydrolase, alpha-D-xylopyranose-(1-6)-beta-D-glucopyranose-(1-4)-[alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-[alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Yaoi, K, Kondo, H, Hiyoshi, A, Noro, N, Sugimoto, H, Miyazaki, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-02-09 | | Release date: | 2007-06-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Structural Basis for the Exo-mode of Action in GH74 Oligoxyloglucan Reducing End-specific Cellobiohydrolase.
J.Mol.Biol., 370, 2007
|
|
