6LGO
 
 | |
6LD9
 
 | |
6LD7
 
 | |
3DKU
 
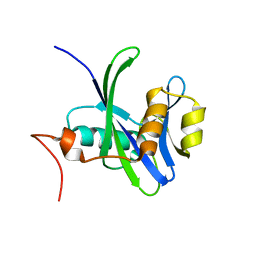 | | Crystal structure of Nudix hydrolase Orf153, ymfB, from Escherichia coli K-1 | | Descriptor: | Putative phosphohydrolase | | Authors: | Hong, M.K, Kim, J.K, Jung, J.H, Jung, J.W, Choi, J.Y, Kang, L.W. | | Deposit date: | 2008-06-26 | | Release date: | 2009-06-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Crystal structure of Nudix hydrolase Orf153, ymfB, from Escherichia coli K-1.
To be Published
|
|
3E5N
 
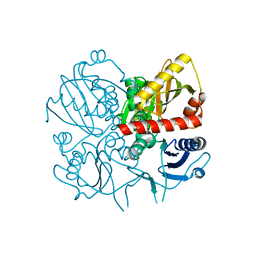 | | Crystal structure of D-alanine-D-alanine ligase from Xanthomonas oryzae pv. oryzae KACC10331 | | Descriptor: | D-alanine-D-alanine ligase A | | Authors: | Doan, T.N.T, Kim, J.K, Kim, H.S, Ahn, Y.J, Kim, J.G, Lee, B.M, Kang, L.W. | | Deposit date: | 2008-08-14 | | Release date: | 2009-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of D-alanine-D-alanine ligase from Xanthomonas oryzae pv. oryzae KACC10331
To be published
|
|
3FK5
 
 | |
4QD4
 
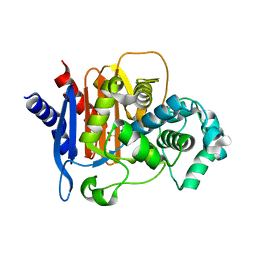 | | Structure of ADC-68, a Novel Carbapenem-Hydrolyzing Class C Extended-Spectrum -Lactamase from Acinetobacter baumannii | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-lactamase ADC-68, CITRIC ACID | | Authors: | Hong, M.K, Kang, L.W. | | Deposit date: | 2014-05-13 | | Release date: | 2015-01-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of ADC-68, a novel carbapenem-hydrolyzing class C extended-spectrum beta-lactamase isolated from Acinetobacter baumannii
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4QF6
 
 | |
4NFW
 
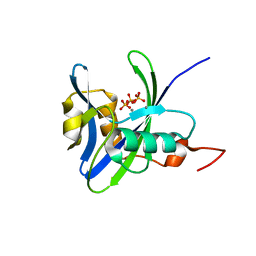 | | Structure and atypical hydrolysis mechanism of the Nudix hydrolase Orf153 (YmfB) from Escherichia coli | | Descriptor: | MANGANESE (II) ION, Putative Nudix hydrolase ymfB, SULFATE ION | | Authors: | Hong, M.K, Kim, J.K, Kang, L.W. | | Deposit date: | 2013-11-01 | | Release date: | 2014-05-14 | | Last modified: | 2015-03-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Divalent metal ion-based catalytic mechanism of the Nudix hydrolase Orf153 (YmfB) from Escherichia coli
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4REJ
 
 | |
4REI
 
 | |
4NT8
 
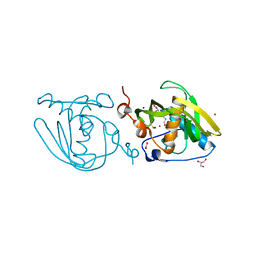 | | Formyl-methionine-alanine complex structure of peptide deformylase from Xanthomoonas oryzae pv. oryzae | | Descriptor: | ACETATE ION, ALANINE, CADMIUM ION, ... | | Authors: | Ngo, H.P.T, Kim, J.K, Kang, L.W. | | Deposit date: | 2013-12-02 | | Release date: | 2014-12-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Substrate complex structure of Xoo1075, a peptide deformylase, from Xanthomonas oryzae pv. oryzae
To be Published
|
|
4QET
 
 | |
4REH
 
 | |
5CP0
 
 | |
5CPD
 
 | |
5BPF
 
 | | Crystal structure of ADP complexed D-alanine-D-alanine ligase(DDL) from Yersinia pestis | | Descriptor: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, D-alanine-D-alanine ligase, ... | | Authors: | Tran, H.T, Kang, L.W, Hong, M.K. | | Deposit date: | 2015-05-28 | | Release date: | 2016-03-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structure of D-alanine-D-alanine ligase from Yersinia pestis: nucleotide phosphate recognition by the serine loop.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
3JRU
 
 | | Crystal structure of Leucyl Aminopeptidase (pepA) from Xoo0834,Xanthomonas oryzae pv. oryzae KACC10331 | | Descriptor: | CALCIUM ION, CARBONATE ION, Probable cytosol aminopeptidase, ... | | Authors: | Natarajan, S, Huynh, K.-H, Kang, L.W. | | Deposit date: | 2009-09-08 | | Release date: | 2010-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of Leucyl Aminopeptidase (pepA) from Xoo0834,Xanthomonas oryzae pv. oryzae KACC10331
to be published
|
|
3K89
 
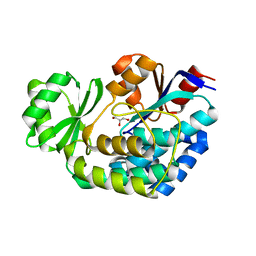 | |
5GK5
 
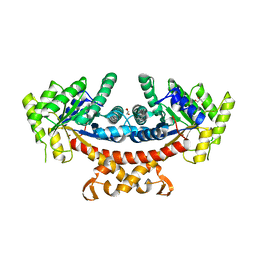 | | Apo structure of fructose 1,6-bisphosphate aldolase from Escherichia coli at 1.9 angstrom resolution | | Descriptor: | DI(HYDROXYETHYL)ETHER, Fructose-bisphosphate aldolase class 2, GLYCEROL, ... | | Authors: | Tran, T.H, Huynh, K.H, Ho, T.H, Kang, L.W. | | Deposit date: | 2016-07-03 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Apo structure of fructose 1,6-bisphosphate aldolase from Escherichia coli at 1.9 angstrom resolution
To Be Published
|
|
5GK3
 
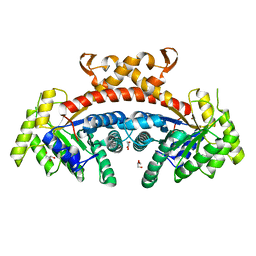 | | Native structure of fructose 1,6-bisphosphate aldolase from Escherichia coli at 1.8 Angstrom resolution | | Descriptor: | DI(HYDROXYETHYL)ETHER, Fructose-bisphosphate aldolase class 2, GLYCEROL, ... | | Authors: | Tran, T.H, Huynh, K.H, Ho, T.H, Kang, L.W. | | Deposit date: | 2016-07-03 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Apo structure of fructose 1,6-bisphosphate aldolase from Escherichia coli at 1.8 Angstrom resolution
To Be Published
|
|
5GK6
 
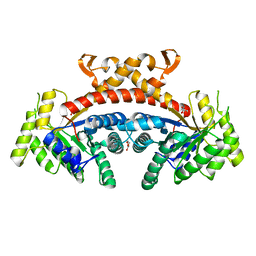 | | Structure of E.Coli fructose 1,6-bisphosphate aldolase, Citrate bound form | | Descriptor: | CITRIC ACID, DI(HYDROXYETHYL)ETHER, Fructose-bisphosphate aldolase class 2, ... | | Authors: | Tran, T.H, Huynh, K.H, Ho, T.H, Kang, L.W. | | Deposit date: | 2016-07-03 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of E.Coli fructose 1,6-bisphosphate aldolase, Citrate bound form
To Be Published
|
|
5GK8
 
 | | Structure of E.Coli fructose 1,6-bisphosphate aldolase, Acetate bound form | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, Fructose-bisphosphate aldolase class 2, ... | | Authors: | Tran, T.H, Huynh, K.H, Ho, T.H, Kang, L.W. | | Deposit date: | 2016-07-03 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Structure of E.Coli fructose 1,6-bisphosphate aldolase, Acetate bound form
To Be Published
|
|
5GT6
 
 | | Apo structure of Aldehyde Dehydrogenase from Bacillus cereus | | Descriptor: | Betaine-aldehyde dehydrogenase, SODIUM ION | | Authors: | Ngo, H.P.T, Hong, S.H, Ho, T.H, Oh, D.K, Kang, L.W. | | Deposit date: | 2016-08-18 | | Release date: | 2017-09-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | crystal structures of aldehyde dehydrogenase from Bacillus cereus having atypical bidirectional oxidizing and reducing activities for all-trans-retinal
To Be Published
|
|
5GK7
 
 | | Structure of E.Coli fructose 1,6-bisphosphate aldolase bound to Tris | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, Fructose-bisphosphate aldolase class 2, ... | | Authors: | Tran, T.H, Huynh, K.H, Ho, T.H, Kang, L.W. | | Deposit date: | 2016-07-03 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of E.Coli fructose 1,6-bisphosphate aldolase, Tris bound form
To Be Published
|
|
