5F06
 
 | | Crystal structure of glutathione transferase F7 from Populus trichocarpa | | Descriptor: | GLUTATHIONE, Glutathione S-transferase family protein, SULFATE ION | | Authors: | Didierjean, C, Rouhier, N, Pegeot, H, Gense, F. | | Deposit date: | 2015-11-27 | | Release date: | 2016-12-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural plasticity among glutathione transferase Phi members: natural combination of catalytic residues confers dual biochemical activities.
FEBS J., 284, 2017
|
|
3PPU
 
 | |
8A0O
 
 | |
8A0I
 
 | |
8A08
 
 | |
7ZVP
 
 | |
7ZS3
 
 | | Crystal structure of poplar glutathione transferase U19 | | Descriptor: | ACETATE ION, Glutathione transferase | | Authors: | Didierjean, C, Favier, F. | | Deposit date: | 2022-05-06 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.603 Å) | | Cite: | Biochemical and Structural Insights on the Poplar Tau Glutathione Transferase GSTU19 and 20 Paralogs Binding Flavonoids.
Front Mol Biosci, 9, 2022
|
|
8A0R
 
 | |
8A0P
 
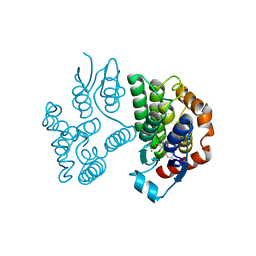 | | Crystal structure of poplar glutathione transferase U20 in complex with morin | | Descriptor: | 2-[2,4-bis(oxidanyl)phenyl]-3,5,7-tris(oxidanyl)chromen-4-one, CHLORIDE ION, Glutathione transferase | | Authors: | Didierjean, C, Favier, F. | | Deposit date: | 2022-05-30 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.686 Å) | | Cite: | Biochemical and Structural Insights on the Poplar Tau Glutathione Transferase GSTU19 and 20 Paralogs Binding Flavonoids.
Front Mol Biosci, 9, 2022
|
|
8A0Q
 
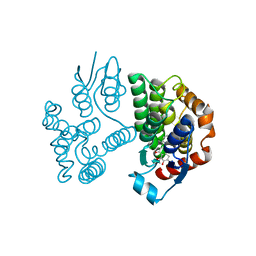 | |
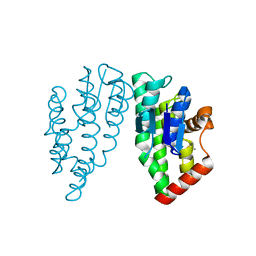
7ZZN
 
 | |
8PFE
 
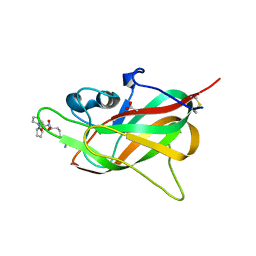 | |
1T90
 
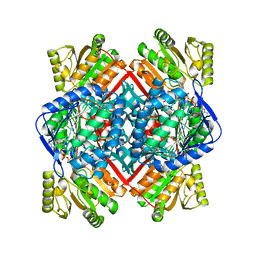 | | Crystal structure of methylmalonate semialdehyde dehydrogenase from Bacillus subtilis | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Probable methylmalonate-semialdehyde dehydrogenase | | Authors: | Dubourg, H, Didierjean, C, Stines-Chaumeil, C, Talfournier, F, Branlant, G, Aubry, A, Corbier, C. | | Deposit date: | 2004-05-14 | | Release date: | 2006-01-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure analysis of Methylmalonate-Semialdehyde Dehydrogenase from Bacillus subtilis.
To be published
|
|
4USS
 
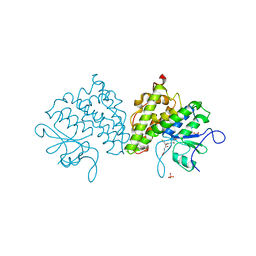 | | Populus trichocarpa glutathione transferase X1-1 (GHR1), complexed with glutathione | | Descriptor: | GLUTATHIONE, GLUTATHIONYL HYDROQUINONE REDUCTASE, PHOSPHATE ION | | Authors: | Lallement, P.A, Meux, E, Gualberto, J.M, Dumaracay, S, Favier, F, Didierjean, C, Saul, F, Haouz, A, Morel-Rouhier, M, Gelhaye, E, Rouhier, N, Hecker, A. | | Deposit date: | 2014-07-13 | | Release date: | 2014-12-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Glutathionyl-Hydroquinone Reductases from Poplar are Plastidial Proteins that Deglutathionylate Both Reduced and Oxidized Glutathionylated Quinones.
FEBS Lett., 589, 2015
|
|
2XF8
 
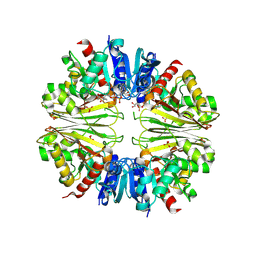 | | Structure of the D-Erythrose-4-Phosphate Dehydrogenase from E. coli in complex with a NAD cofactor analog (3-Chloroacetyl adenine pyridine dinucleotide) and sulfate anion | | Descriptor: | 3-(CHLOROACETYL) PYRIDINE ADENINE DINUCLEOTIDE, D-ERYTHROSE-4-PHOSPHATE DEHYDROGENASE, SULFATE ION | | Authors: | Moniot, S, Didierjean, C, Boschi-Muller, S, Branlant, G, Corbier, C. | | Deposit date: | 2010-05-20 | | Release date: | 2011-06-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural Characterization of Erythrose-4- Phosphate Dehydrogenase from Escherichia Coli: Peculiar Features When Compared to Phosphorylating Gapdhs
To be Published
|
|
6ZGN
 
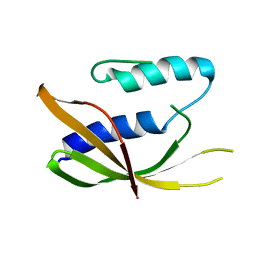 | | Crystal structure of VirB8-like OrfG central domain of Streptococcus thermophilus ICESt3; a putative assembly factor of a gram positive conjugative Type IV secretion system. | | Descriptor: | Putative transfer protein | | Authors: | Cappele, J, Mohamad-Ali, A, Leblond-Bourget, N, Payot-Lacroix, S, Mathiot, S, Didierjean, C, Favier, F, Douzi, B. | | Deposit date: | 2020-06-19 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and Biochemical Analysis of OrfG: The VirB8-like Component of the Conjugative Type IV Secretion System of ICE St3 From Streptococcus thermophilus .
Front Mol Biosci, 8, 2021
|
|
2ESD
 
 | | Crystal Structure of thioacylenzyme intermediate of an Nadp Dependent Aldehyde Dehydrogenase | | Descriptor: | GLYCERALDEHYDE-3-PHOSPHATE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADP-dependent glyceraldehyde-3-phosphate dehydrogenase | | Authors: | D'Ambrosio, K, Didierjean, C, Benedetti, E, Aubry, A, Corbier, C. | | Deposit date: | 2005-10-26 | | Release date: | 2006-05-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The first crystal structure of a thioacylenzyme intermediate in the ALDH family: new coenzyme conformation and relevance to catalysis
Biochemistry, 45, 2006
|
|
2X5J
 
 | | Crystal structure of the Apoform of the D-Erythrose-4-phosphate dehydrogenase from E. coli | | Descriptor: | D-ERYTHROSE-4-PHOSPHATE DEHYDROGENASE, PHOSPHATE ION | | Authors: | Moniot, S, Didierjean, C, Boschi-Muller, S, Branlant, G, Corbier, C. | | Deposit date: | 2010-02-09 | | Release date: | 2011-02-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Characterization of Erythrose-4- Phosphate Dehydrogenase from Escherichia Coli: Peculiar Features When Compared to Phosphorylating Gapdhs
To be Published
|
|
2X5K
 
 | | Structure of an active site mutant of the D-Erythrose-4-Phosphate Dehydrogenase from E. coli | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, D-ERYTHROSE-4-PHOSPHATE DEHYDROGENASE, ... | | Authors: | Moniot, S, Didierjean, C, Boschi-Muller, S, Branlant, G, Corbier, C. | | Deposit date: | 2010-02-10 | | Release date: | 2011-02-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structural Characterization of Erythrose-4- Phosphate Dehydrogenase from Escherichia Coli: Peculiar Features When Compared to Phosphorylating Gapdhs
To be Published
|
|
7PKW
 
 | | Crystal structure of VIRB8-like OrfG central and C-terminal domains of Streptococcus thermophilus ICESt3 (Gram positive conjugative type IV secretion system). | | Descriptor: | GLYCEROL, Putative transfer protein, SULFATE ION | | Authors: | Favier, F, Didierjean, C, Cappele, J, Douzi, B, Leblond-Bourget, N. | | Deposit date: | 2021-08-27 | | Release date: | 2022-09-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.835 Å) | | Cite: | Crystal structure of VIRB8-like OrfG central and C-terminal domains of Streptococcus thermophilus ICESt3 (Gram positive conjugative type IV secretion system).
To Be Published
|
|
3CMC
 
 | | Thioacylenzyme intermediate of Bacillus stearothermophilus phosphorylating GAPDH | | Descriptor: | 1,2-ETHANEDIOL, GLYCERALDEHYDE-3-PHOSPHATE, GLYCEROL, ... | | Authors: | Moniot, S, Vonrhein, C, Bricogne, G, Didierjean, C, Corbier, C. | | Deposit date: | 2008-03-21 | | Release date: | 2008-06-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Trapping of the Thioacylglyceraldehyde-3-phosphate Dehydrogenase Intermediate from Bacillus stearothermophilus: DIRECT EVIDENCE FOR A FLIP-FLOP MECHANISM
J.Biol.Chem., 283, 2008
|
|
5LKD
 
 | | Crystal structure of the Xi glutathione transferase ECM4 from Saccharomyces cerevisiae in complex with glutathione | | Descriptor: | GLUTATHIONE, Glutathione S-transferase omega-like 2 | | Authors: | Schwartz, M, Didierjean, C, Hecker, A, Girardet, J.M, Morel-Rouhier, M, Gelhaye, E, Favier, F. | | Deposit date: | 2016-07-22 | | Release date: | 2016-10-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal Structure of Saccharomyces cerevisiae ECM4, a Xi-Class Glutathione Transferase that Reacts with Glutathionyl-(hydro)quinones.
Plos One, 11, 2016
|
|
5LKB
 
 | | Crystal structure of the Xi glutathione transferase ECM4 from Saccharomyces cerevisiae | | Descriptor: | GLYCEROL, Glutathione S-transferase omega-like 2 | | Authors: | Schwartz, M, Didierjean, C, Hecker, A, Girardet, J.M, Morel-Rouhier, M, Gelhaye, E, Favier, F. | | Deposit date: | 2016-07-22 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal Structure of Saccharomyces cerevisiae ECM4, a Xi-Class Glutathione Transferase that Reacts with Glutathionyl-(hydro)quinones.
Plos One, 11, 2016
|
|
4G10
 
 | | LigG from Sphingobium sp. SYK-6 is related to the glutathione transferase omega class | | Descriptor: | ACETATE ION, GLUTATHIONE, Glutathione S-transferase homolog, ... | | Authors: | Meux, E, Prosper, P, Masai, E, Mulliert Carlin, G, Dumarcay, S, Morel, M, Didierjean, C, Gelhaye, E, Favier, F. | | Deposit date: | 2012-07-10 | | Release date: | 2012-10-03 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Sphingobium sp. SYK-6 LigG involved in lignin degradation is structurally and biochemically related to the glutathione transferase omega class.
Febs Lett., 586, 2012
|
|
7NCW
 
 | |
