8DOQ
 
 | |
8DQ9
 
 | |
8FI3
 
 | |
8FUY
 
 | |
8G0V
 
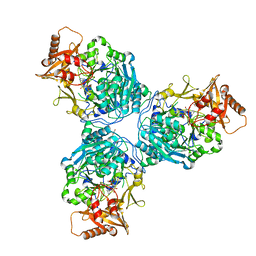 | |
8G0T
 
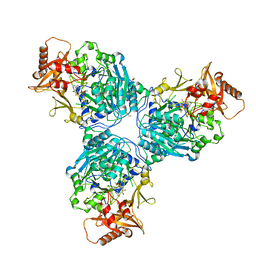 | |
8G0U
 
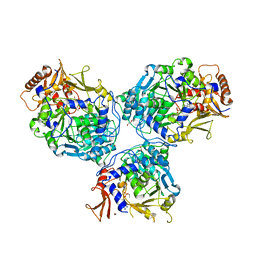 | |
8F8U
 
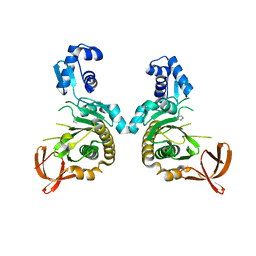 | |
8G0S
 
 | |
8U9A
 
 | | Crystal Structure of 2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase from Klebsiella aerogenes (DBH bound) | | Descriptor: | 2,3-DIHYDROXY-BENZOIC ACID, 2,3-dihydroxybenzoate-2,3-dehydrogenase, CHLORIDE ION | | Authors: | Seattle Structural Genomics Center for Infectious Disease, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2023-09-18 | | Release date: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of 2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase from Klebsiella aerogenes (DBH bound)
To be published
|
|
7TM9
 
 | |
7TM4
 
 | |
7TM6
 
 | |
7TM7
 
 | |
7TM5
 
 | |
7TM8
 
 | |
7TMV
 
 | |
4MO7
 
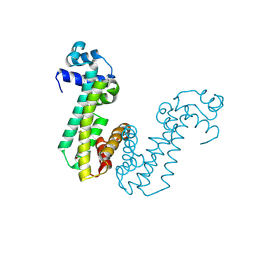 | | Crystal structure of superantigen PfiT | | Descriptor: | BETA-MERCAPTOETHANOL, MAGNESIUM ION, Transcriptional regulator I2 | | Authors: | Liu, L.H, Chen, H, Li, H.M. | | Deposit date: | 2013-09-11 | | Release date: | 2014-02-05 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Pfit is a structurally novel Crohn's disease-associated superantigen.
Plos Pathog., 9, 2013
|
|
4MXM
 
 | |
8F87
 
 | |
8F82
 
 | |
8F81
 
 | |
8F80
 
 | |
8SBY
 
 | | Crystal Structure of 2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase from Klebsiella aerogenes (NAD and sulfate bound, hexagonal form) | | Descriptor: | 2,3-dihydroxybenzoate-2,3-dehydrogenase, CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2023-04-04 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of 2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase from Klebsiella aerogenes (NAD and sulfate bound, hexagonal form)
To be published
|
|
8SBX
 
 | | Crystal Structure of 2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase from Klebsiella aerogenes (Apo, hexagonal form) | | Descriptor: | 2,3-dihydroxybenzoate-2,3-dehydrogenase, CHLORIDE ION, SULFATE ION | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2023-04-04 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of 2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase from Klebsiella aerogenes (Apo, hexagonal form)
To be published
|
|
