1TT3
 
 | | NMR soulution structure of omega-conotoxin [K10]MVIIA | | Descriptor: | Omega-conotoxin MVIIa | | Authors: | Adams, D.J, Smith, A.B, Schroeder, C.I, Yasuda, T, Lewis, R.J. | | Deposit date: | 2004-06-21 | | Release date: | 2004-07-06 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | omega-conotoxin CVID inhibits a pharmacologically distinct voltage-sensitive calcium channel associated with transmitter release from preganglionic nerve terminals
J.Biol.Chem., 278, 2003
|
|
1TU6
 
 | | Cathepsin K complexed with a ketoamide inhibitor | | Descriptor: | Cathepsin K, SULFATE ION, [1-(4-FLUOROBENZYL)CYCLOBUTYL]METHYL (1S)-1-[OXO(1H-PYRAZOL-5-YLAMINO)ACETYL]PENTYLCARBAMATE | | Authors: | Barrett, D.G, Catalano, J.G, Deaton, D.N, Hassell, A.M, Long, S.T, Miller, A.B, Miller, L.R, Shewchuk, L.M, Wells-Knecht, K.J, Wright, L.L. | | Deposit date: | 2004-06-24 | | Release date: | 2004-09-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Potent and selective P2-P3 ketoamide inhibitors of cathepsin K with improved pharmacokinetic properties via favorable P1', P1, and/or P3 substitutions
BIOORG.MED.CHEM.LETT., 14, 2004
|
|
1UB6
 
 | | Crystal structure of Antibody 19G2 with sera ligand | | Descriptor: | antibody 19G2, alpha chain, beta chain | | Authors: | Beuscher, A.B, Wirsching, P, Lerner, R.A, Janda, K, Stevens, R.C. | | Deposit date: | 2003-03-30 | | Release date: | 2004-04-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structure and Dynamics of Blue Fluorescent Antibody 19G2 at Blue and Violet Fluorescent Temperatures
To be published
|
|
1W9T
 
 | | Structure of a beta-1,3-glucan binding CBM6 from Bacillus halodurans in complex with xylobiose | | Descriptor: | BH0236 PROTEIN, SODIUM ION, alpha-D-xylopyranose, ... | | Authors: | Boraston, A.B, van Bueren, A.L. | | Deposit date: | 2004-10-18 | | Release date: | 2004-11-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Family 6 Carbohydrate Binding Modules Recognize the Non-Reducing End of Beta-1,3-Linked Glucans by Presenting a Unique Ligand Binding Surface
J.Biol.Chem., 280, 2005
|
|
1W9S
 
 | |
1B2W
 
 | | COMPARISON OF THE THREE-DIMENSIONAL STRUCTURES OF A HUMANIZED AND A CHIMERIC FAB OF AN ANTI-GAMMA-INTERFERON ANTIBODY | | Descriptor: | PROTEIN (ANTIBODY (HEAVY CHAIN)), PROTEIN (ANTIBODY (LIGHT CHAIN)) | | Authors: | Fan, Z, Shan, L, Goldsteen, B.Z, Guddat, L.W, Thakur, A, Landolfi, N.F, Co, M.S, Vasquez, M, Queen, C, Ramsland, P.A, Edmundson, A.B. | | Deposit date: | 1998-12-01 | | Release date: | 1999-05-06 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Comparison of the three-dimensional structures of a humanized and a chimeric Fab of an anti-gamma-interferon antibody.
J.Mol.Recog., 12, 1999
|
|
1B4J
 
 | | COMPARISON OF THE THREE-DIMENSIONAL STRUCTURES OF A HUMANIZED AND A CHIMERIC FAB OF AN ANTI-GAMMA-INTERFERON ANTIBODY | | Descriptor: | ANTIBODY | | Authors: | Fan, Z, Shan, L, Goldsteen, B.Z, Guddat, L.W, Thakur, A, Landolfi, N.F, Co, M.S, Vasques, M, Queen, C, Ramsland, P.A, Edmundson, A.B. | | Deposit date: | 1998-12-22 | | Release date: | 1999-06-15 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Comparison of the three-dimensional structures of a humanized and a chimeric Fab of an anti-gamma-interferon antibody.
J.Mol.Recog., 12, 1999
|
|
1AQK
 
 | |
1WBH
 
 | |
1W9W
 
 | |
1WA3
 
 | | Mechanism of the Class I KDPG aldolase | | Descriptor: | 2-KETO-3-DEOXY-6-PHOSPHOGLUCONATE ALDOLASE, PYRUVIC ACID, SULFATE ION | | Authors: | Fullerton, S.W.B, Griffiths, J.S, Merkel, A.B, Wymer, N.J, Hutchins, M.J, Fierke, C.A, Toone, E.J, Naismith, J.H. | | Deposit date: | 2004-10-22 | | Release date: | 2005-01-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanism of the Class I Kdpg Aldolase.
Bioorg.Med.Chem., 14, 2006
|
|
1WAU
 
 | |
1W0N
 
 | | Structure of uncomplexed Carbohydrate Binding Domain CBM36 | | Descriptor: | CALCIUM ION, ENDO-1,4-BETA-XYLANASE D, MAGNESIUM ION, ... | | Authors: | Jamal, S, Boraston, A.B, Davies, G.J. | | Deposit date: | 2004-06-09 | | Release date: | 2004-10-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (0.8 Å) | | Cite: | Ab Initio Structure Determination and Functional Characterization of Cbm36: A New Family of Calcium-Dependent Carbohydrate Binding Modules
Structure, 12, 2004
|
|
1UX7
 
 | | Carbohydrate-Binding Module CBM36 in complex with calcium and xylotriose | | Descriptor: | CALCIUM ION, ENDO-1,4-BETA-XYLANASE D, SULFATE ION, ... | | Authors: | Davies, G.J, Boraston, A.B, Jamal, S. | | Deposit date: | 2004-02-19 | | Release date: | 2004-10-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Ab Initio Structure Determination and Functional Characterization of Cbm36: A New Family of Calcium-Dependent Carbohydrate Binding Modules
Structure, 12, 2004
|
|
1T66
 
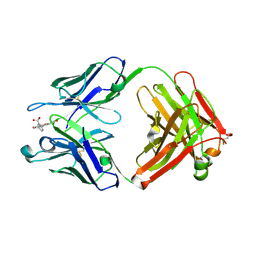 | | The structure of FAB with intermediate affinity for fluorescein. | | Descriptor: | 2-(6-HYDROXY-3-OXO-3H-XANTHEN-9-YL)-BENZOIC ACID, immunoglobulin heavy chain, immunoglobulin light chain | | Authors: | Terzyan, S, Ramsland, P.A, Voss Jr, E.W, Herron, J.N, Edmundson, A.B. | | Deposit date: | 2004-05-05 | | Release date: | 2004-05-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Three-dimensional Structures of Idiotypically Related Fabs with Intermediate and High Affinity for Fluorescein.
J.Mol.Biol., 339, 2004
|
|
1A8F
 
 | | HUMAN SERUM TRANSFERRIN, RECOMBINANT N-TERMINAL LOBE | | Descriptor: | CARBONATE ION, FE (III) ION, SERUM TRANSFERRIN | | Authors: | Macgillivray, R.T.A, Moore, S.A, Chen, J, Anderson, B.F, Baker, H, Luo, Y, Bewley, M, Smith, C.A, Murphy, M.E.P, Wang, Y, Mason, A.B, Woodworth, R.C, Brayer, G.D, Baker, E.N. | | Deposit date: | 1998-03-25 | | Release date: | 1998-06-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Two high-resolution crystal structures of the recombinant N-lobe of human transferrin reveal a structural change implicated in iron release.
Biochemistry, 37, 1998
|
|
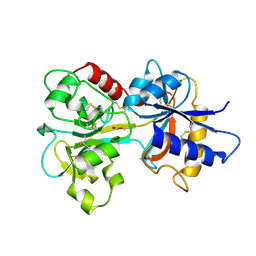
1A8P
 
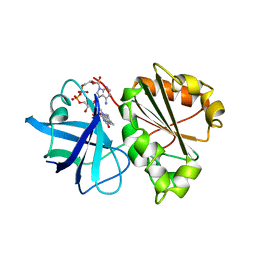 | | FERREDOXIN REDUCTASE FROM AZOTOBACTER VINELANDII | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADPH:FERREDOXIN OXIDOREDUCTASE | | Authors: | Prasad, G.S, Kresge, N, Muhlberg, A.B, Shaw, A, Jung, Y.S, Burgess, B.K, Stout, C.D. | | Deposit date: | 1998-03-28 | | Release date: | 1998-06-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of NADPH:ferredoxin reductase from Azotobacter vinelandii.
Protein Sci., 7, 1998
|
|
1B6B
 
 | |
1B3E
 
 | | HUMAN SERUM TRANSFERRIN, N-TERMINAL LOBE, EXPRESSED IN PICHIA PASTORIS | | Descriptor: | CARBONATE ION, FE (III) ION, PROTEIN (SERUM TRANSFERRIN) | | Authors: | Bewley, M.C, Tam, B.M, Grewal, J, He, S, Shewry, S, Murphy, M.E.P, Mason, A.B, Woodworth, R.C, Baker, E.N, Macgillivray, R.T.A. | | Deposit date: | 1998-12-09 | | Release date: | 1999-03-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray crystallography and mass spectroscopy reveal that the N-lobe of human transferrin expressed in Pichia pastoris is folded correctly but is glycosylated on serine-32.
Biochemistry, 38, 1999
|
|
1UY4
 
 | |
1UY3
 
 | |
1UY1
 
 | |
1SZP
 
 | | A Crystal Structure of the Rad51 Filament | | Descriptor: | DNA repair protein RAD51, SULFATE ION | | Authors: | Conway, A.B, Lynch, T.W, Zhang, Y, Fortin, G.S, Symington, L.S, Rice, P.A. | | Deposit date: | 2004-04-06 | | Release date: | 2004-07-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Crystal structure of a Rad51 filament.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1T8I
 
 | | Human DNA Topoisomerase I (70 Kda) In Complex With The Poison Camptothecin and Covalent Complex With A 22 Base Pair DNA Duplex | | Descriptor: | 4-ETHYL-4-HYDROXY-1,12-DIHYDRO-4H-2-OXA-6,12A-DIAZA-DIBENZO[B,H]FLUORENE-3,13-DIONE, 5'-D(*(TGP)P*GP*AP*AP*AP*AP*AP*TP*TP*TP*TP*T)-3', 5'-D(*AP*AP*AP*AP*AP*GP*AP*CP*TP*T)-3', ... | | Authors: | Staker, B.L, Feese, M.D, Cushman, M, Pommier, Y, Zembower, D, Stewart, L, Burgin, A.B. | | Deposit date: | 2004-05-12 | | Release date: | 2005-05-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of three classes of anticancer agents bound to the human topoisomerase I-DNA covalent complex
J.Med.Chem., 48, 2005
|
|
1A8C
 
 | | PRIMARY SEQUENCE AND SOLUTION CONFORMATION OF FERROCYTOCHROME C-552 FROM NITROSOMONAS EUROPAEA, NMR, MEAN STRUCTURE REFINED WITHOUT HYDROGEN BOND CONSTRAINTS | | Descriptor: | FERROCYTOCHROME C-552, HEME C | | Authors: | Timkovich, R, Bergmann, D, Arciero, D.M, Hooper, A.B. | | Deposit date: | 1998-03-23 | | Release date: | 1998-10-21 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Primary sequence and solution conformation of ferrocytochrome c-552 from Nitrosomonas europaea.
Biophys.J., 75, 1998
|
|
