1IC2
 
 | | DECIPHERING THE DESIGN OF THE TROPOMYOSIN MOLECULE | | Descriptor: | TROPOMYOSIN ALPHA CHAIN, SKELETAL MUSCLE | | Authors: | Brown, J.H, Kim, K.-H, Jun, G, Greenfield, N.J, Dominguez, R, Volkmann, N, Hitchcock-DeGregori, S.E, Cohen, C. | | Deposit date: | 2001-03-29 | | Release date: | 2001-07-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Deciphering the design of the tropomyosin molecule
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
2A13
 
 | | X-ray structure of protein from Arabidopsis thaliana AT1G79260 | | Descriptor: | At1g79260 | | Authors: | Wesenberg, G.E, Phillips Jr, G.N, McCoy, J.G, Bitto, E, Bingman, C.A, Allard, S.T.M, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2005-06-17 | | Release date: | 2005-07-12 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | The structure and NO binding properties of the nitrophorin-like heme-binding protein from Arabidopsis thaliana gene locus At1g79260.1.
Proteins, 78, 2010
|
|
2ATF
 
 | | X-RAY STRUCTURE OF cysteine dioxygenase type I FROM MUS MUSCULUS MM.241056 | | Descriptor: | 1,2-ETHANEDIOL, Cysteine dioxygenase type I, NICKEL (II) ION | | Authors: | Wesenberg, G.E, Phillips Jr, G.N, Mccoy, J.G, Bitto, E, Bingman, C.A, Allard, S.T.M, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2005-08-24 | | Release date: | 2005-10-18 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and mechanism of mouse cysteine dioxygenase.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2APJ
 
 | | X-Ray Structure of Protein from Arabidopsis Thaliana AT4G34215 at 1.6 Angstrom Resolution | | Descriptor: | Putative Esterase | | Authors: | Wesenberg, G.E, Phillips Jr, G.N, Mccoy, J.G, Bitto, E, Bingman, C.A, Allard, S.T, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2005-08-16 | | Release date: | 2005-08-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structure at 1.6 Angstroms resolution of the protein product of the At4g34215 gene from Arabidopsis thaliana.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2BDU
 
 | | X-Ray Structure of a Cytosolic 5'-Nucleotidase III from Mus Musculus MM.158936 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Cytosolic 5'-nucleotidase III | | Authors: | Wesenberg, G.E, Phillips Jr, G.N, Han, B.W, Bitto, E, Bingman, C.A, Bae, E, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2005-10-20 | | Release date: | 2005-11-01 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of pyrimidine 5'-nucleotidase type 1. Insight into mechanism of action and inhibition during lead poisoning.
J.Biol.Chem., 281, 2006
|
|
2BEI
 
 | | X-ray structure of thialysine n-acetyltransferase (SSAT2) from homo sapiens | | Descriptor: | ACETYL COENZYME *A, Diamine acetyltransferase 2 | | Authors: | Wesenberg, G.E, Phillips Jr, G.N, Han, B.W, Bitto, E, Bingman, C.A, Bae, E, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2005-10-24 | | Release date: | 2005-11-01 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.842 Å) | | Cite: | Crystal structure of Homo sapiens thialysine Nepsilon-acetyltransferase (HsSSAT2) in complex with acetyl coenzyme A.
Proteins, 64, 2006
|
|
1VJI
 
 | | Gene Product of At1g76680 from Arabidopsis thaliana | | Descriptor: | 12-oxophytodienoate reductase (OPR1), FLAVIN MONONUCLEOTIDE | | Authors: | Wesenberg, G.E, Smith, D.W, Phillips Jr, G.N, Johnson, K.A, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-02-24 | | Release date: | 2004-03-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | X-ray structure of Arabidopsis At1g77680, 12-oxophytodienoate reductase isoform 1.
Proteins, 61, 2005
|
|
2EXR
 
 | | X-Ray Structure of Cytokinin Oxidase/Dehydrogenase (CKX) From Arabidopsis Thaliana AT5G21482 | | Descriptor: | Cytokinin dehydrogenase 7, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Wesenberg, G.E, Phillips Jr, G.N, Han, B.W, Bitto, E, Bingman, C.A, Bae, E, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2005-11-08 | | Release date: | 2005-11-29 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Crystal structure of Arabidopsis thaliana cytokinin dehydrogenase.
Proteins, 70, 2008
|
|
2G07
 
 | | X-ray structure of mouse pyrimidine 5'-nucleotidase type 1, phospho-enzyme intermediate analog with Beryllium fluoride | | Descriptor: | Cytosolic 5'-nucleotidase III, MAGNESIUM ION | | Authors: | Bitto, E, Bingman, C.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-02-11 | | Release date: | 2006-04-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of pyrimidine 5'-nucleotidase type 1. Insight into mechanism of action and inhibition during lead poisoning.
J.Biol.Chem., 281, 2006
|
|
2G06
 
 | | X-ray structure of mouse pyrimidine 5'-nucleotidase type 1, with bound magnesium(II) | | Descriptor: | Cytosolic 5'-nucleotidase III, MAGNESIUM ION, PIPERAZINE-N,N'-BIS(2-ETHANESULFONIC ACID) | | Authors: | Bitto, E, Bingman, C.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-02-11 | | Release date: | 2006-04-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of pyrimidine 5'-nucleotidase type 1. Insight into mechanism of action and inhibition during lead poisoning.
J.Biol.Chem., 281, 2006
|
|
2G0A
 
 | | X-ray structure of mouse pyrimidine 5'-nucleotidase type 1 with lead(II) bound in active site | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Cytosolic 5'-nucleotidase III, LEAD (II) ION | | Authors: | Bitto, E, Bingman, C.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-02-11 | | Release date: | 2006-04-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of pyrimidine 5'-nucleotidase type 1. Insight into mechanism of action and inhibition during lead poisoning.
J.Biol.Chem., 281, 2006
|
|
2G5W
 
 | | X-ray crystal structure of Arabidopsis thaliana 12-oxophytodienoate reductase isoform 3 (AtOPR3) in complex with 8-iso prostaglandin A1 and its cofactor, flavin mononucleotide. | | Descriptor: | (8S,12S)-15S-HYDROXY-9-OXOPROSTA-10Z,13E-DIEN-1-OIC ACID, 12-oxophytodienoate reductase 3, FLAVIN MONONUCLEOTIDE | | Authors: | Han, B.W, Malone, T.E, Bingman, C.A, Wesenberg, G.E, Phillips Jr, G.N, Fox, B.G, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-02-23 | | Release date: | 2006-04-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.576 Å) | | Cite: | Crystal structure of Arabidopsis thaliana 12-oxophytodienoate reductase isoform 3 in complex with 8-iso prostaglandin A(1).
Proteins, 79, 2011
|
|
2G09
 
 | | X-ray structure of mouse pyrimidine 5'-nucleotidase type 1, product complex | | Descriptor: | Cytosolic 5'-nucleotidase III, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Bitto, E, Bingman, C.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-02-11 | | Release date: | 2006-04-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of pyrimidine 5'-nucleotidase type 1. Insight into mechanism of action and inhibition during lead poisoning.
J.Biol.Chem., 281, 2006
|
|
1BAP
 
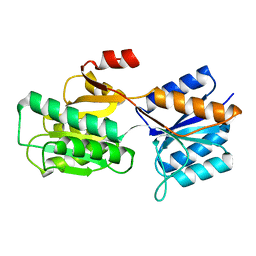 | |
1APB
 
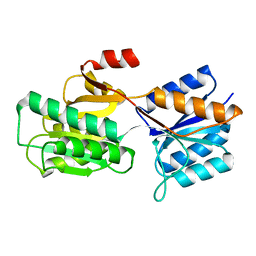 | |
2EVN
 
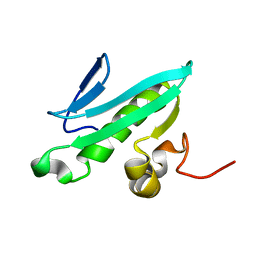 | | NMR solution structures of At1g77540 | | Descriptor: | Protein At1g77540 | | Authors: | Tyler, R.C, Singh, S, Tonelli, M, Min, M.S, Markley, J.L, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2005-10-31 | | Release date: | 2005-11-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of Arabidopsis thaliana At1g77540 Protein, a Minimal Acetyltransferase from the COG2388 Family.
Biochemistry, 45, 2006
|
|
1OR4
 
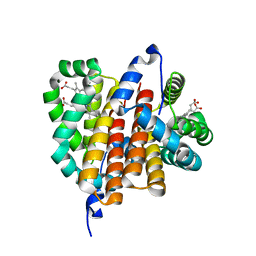 | |
1OR6
 
 | |
1VK5
 
 | | X-ray Structure of Gene Product from Arabidopsis Thaliana At3g22680 | | Descriptor: | 1,2-ETHANEDIOL, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, SULFATE ION, ... | | Authors: | Wesenberg, G.E, Smith, D.W, Phillips Jr, G.N, Johnson, K.A, Bingman, C.A, Allard, S.T.M, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-05-06 | | Release date: | 2004-05-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.604 Å) | | Cite: | Structure at 1.6 A resolution of the protein from gene locus At3g22680 from Arabidopsis thaliana.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
1VM9
 
 | | The X-ray Structure of the cys84ala cys85ala double mutant of the [2Fe-2S] Ferredoxin subunit of Toluene-4-Monooxygenase from Pseudomonas Mendocina KR1 | | Descriptor: | 1,2-ETHANEDIOL, FE2/S2 (INORGANIC) CLUSTER, MAGNESIUM ION, ... | | Authors: | Wesenberg, G.E, Smith, D.W, Phillips Jr, G.N, Bingman, C.A, Allard, S.T.M, Moe, L.A, Fox, B.G, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-09-13 | | Release date: | 2004-09-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structure of T4moC, the Rieske-type ferredoxin component of toluene 4-monooxygenase.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
1XFI
 
 | | X-ray structure of gene product from Arabidopsis thaliana At2g17340 | | Descriptor: | MAGNESIUM ION, unknown protein | | Authors: | Wesenberg, G.E, Smith, D.W, Phillips Jr, G.N, Bitto, E, Bingman, C.A, Allard, S.T.M, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-09-14 | | Release date: | 2004-09-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure at 1.7 A resolution of the protein product of the At2g17340 gene from Arabidopsis thaliana.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
1Y0Z
 
 | | X-ray structure of gene product from Arabidopsis thaliana At3g21360 | | Descriptor: | FE (II) ION, SULFATE ION, protein product of AT3G21360 | | Authors: | Wesenberg, G.E, Phillips Jr, G.N, Bitto, E, Bingman, C.A, Allard, S.T.M, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-11-16 | | Release date: | 2004-11-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structure at 2.4 A resolution of the protein from gene locus At3g21360, a putative Fe(II)/2-oxoglutarate-dependent enzyme from Arabidopsis thaliana.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
1ZTP
 
 | | X-ray structure of gene product from homo sapiens Hs.433573 | | Descriptor: | Basophilic leukemia expressed protein BLES03 | | Authors: | Wesenberg, G.E, Phillips Jr, G.N, Bitto, E, Bingman, C.A, Allard, S.T.M, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2005-05-27 | | Release date: | 2005-06-14 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure at 2.5 A resolution of human basophilic leukemia-expressed protein BLES03.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
2G08
 
 | | X-ray structure of mouse pyrimidine 5'-nucleotidase type 1, product-transition complex analog with Aluminum fluoride | | Descriptor: | ALUMINUM FLUORIDE, Cytosolic 5'-nucleotidase III, MAGNESIUM ION | | Authors: | Bitto, E, Bingman, C.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-02-11 | | Release date: | 2006-04-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of pyrimidine 5'-nucleotidase type 1. Insight into mechanism of action and inhibition during lead poisoning.
J.Biol.Chem., 281, 2006
|
|
2GCU
 
 | | X-Ray Structure of Gene Product from Arabidopsis Thaliana At1g53580 | | Descriptor: | 1,2-ETHANEDIOL, FE (II) ION, Putative hydroxyacylglutathione hydrolase 3, ... | | Authors: | McCoy, J.G, Wesenberg, G.E, Phillips Jr, G.N, Bitto, E, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-03-14 | | Release date: | 2006-04-18 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.477 Å) | | Cite: | Structure of an ETHE1-like protein from Arabidopsis thaliana.
ACTA CRYSTALLOGR.,SECT.D, 62, 2006
|
|
