4FPV
 
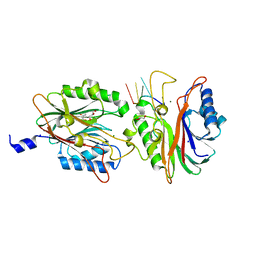 | | Crystal structure of D. rerio TDP2 complexed with single strand DNA product | | Descriptor: | DNA (5'-D(P*TP*GP*CP*AP*G)-3'), GLYCEROL, MAGNESIUM ION, ... | | Authors: | Shi, K, Kurahashi, K, Aihara, H. | | Deposit date: | 2012-06-22 | | Release date: | 2012-10-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural basis for recognition of 5'-phosphotyrosine adducts by Tdp2.
Nat.Struct.Mol.Biol., 19, 2012
|
|
1D0O
 
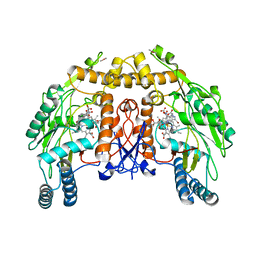 | | BOVINE ENDOTHELIAL NITRIC OXIDE SYNTHASE HEME DOMAIN COMPLEXED WITH 3-BROMO-7-NITROINDAZOLE (H4B PRESENT) | | Descriptor: | 3-BROMO-7-NITROINDAZOLE, ACETATE ION, BOVINE ENDOTHELIAL NITRIC OXIDE SYNTHASE HEME, ... | | Authors: | Raman, C.S, Li, H, Martasek, P, Southan, G.J, Masters, B.S.S, Poulos, T.L. | | Deposit date: | 1999-09-13 | | Release date: | 2001-11-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of nitric oxide synthase bound to nitro indazole reveals a novel inactivation mechanism.
Biochemistry, 40, 2001
|
|
1COB
 
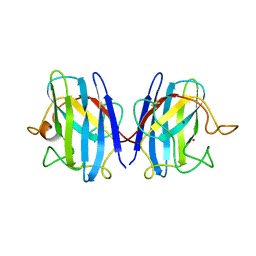 | | CRYSTAL STRUCTURE SOLUTION AND REFINEMENT OF THE SEMISYNTHETIC COBALT SUBSTITUTED BOVINE ERYTHROCYTE ENZYME SUPEROXIDE DISMUTASE AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | COBALT (II) ION, COPPER (II) ION, SUPEROXIDE DISMUTASE | | Authors: | Djinovic, K, Coda, A, Antolini, L, Pelosi, G, Desideri, A, Falconi, M, Rotilio, G, Bolognesi, M. | | Deposit date: | 1992-02-19 | | Release date: | 1993-10-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure solution and refinement of the semisynthetic cobalt-substituted bovine erythrocyte superoxide dismutase at 2.0 A resolution.
J.Mol.Biol., 226, 1992
|
|
4FVA
 
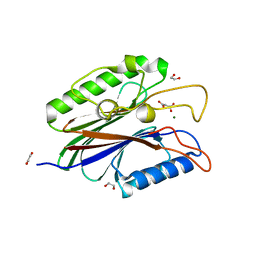 | | Crystal structure of truncated Caenorhabditis elegans TDP2 | | Descriptor: | 1,2-ETHANEDIOL, 5'-tyrosyl-DNA phosphodiesterase, MAGNESIUM ION, ... | | Authors: | Shi, K, Kurahashi, K, Aihara, H. | | Deposit date: | 2012-06-29 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural basis for recognition of 5'-phosphotyrosine adducts by Tdp2.
Nat.Struct.Mol.Biol., 19, 2012
|
|
4GEW
 
 | |
1NSI
 
 | | HUMAN INDUCIBLE NITRIC OXIDE SYNTHASE, ZN-BOUND, L-ARG COMPLEX | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ARGININE, GLYCEROL, ... | | Authors: | Li, H, Raman, C.S, Glaser, C.B, Blasko, E, Young, T.A, Parkinson, J.F, Whitlow, M, Poulos, T.L. | | Deposit date: | 1999-01-10 | | Release date: | 2000-01-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structures of zinc-free and -bound heme domain of human inducible nitric-oxide synthase. Implications for dimer stability and comparison with endothelial nitric-oxide synthase.
J.Biol.Chem., 274, 1999
|
|
1YAZ
 
 | | AZIDE-BOUND YEAST CU(II)/ZN SUPEROXIDE DISMUTASE ROOM TEMPERATURE (298K) STRUCTURE | | Descriptor: | AZIDE ION, COPPER (II) ION, PROTEIN (CU/ZN SUPEROXIDE DISMUTASE), ... | | Authors: | Hart, P.J, Balbirnie, M.M, Ogihara, N.L, Nersissian, A.M, Weiss, M.S, Valentine, J.S, Eisenberg, D. | | Deposit date: | 1998-12-23 | | Release date: | 2000-01-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A structure-based mechanism for copper-zinc superoxide dismutase.
Biochemistry, 38, 1999
|
|
2FBY
 
 | | WRN exonuclease, Eu complex | | Descriptor: | EUROPIUM (III) ION, Werner syndrome helicase | | Authors: | Perry, J.J. | | Deposit date: | 2005-12-10 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | WRN exonuclease structure and molecular mechanism imply an editing role in DNA end processing.
Nat.Struct.Mol.Biol., 13, 2006
|
|
1S8E
 
 | | Crystal structure of Mre11-3 | | Descriptor: | MANGANESE (II) ION, exonuclease putative | | Authors: | Hopfner, K.P. | | Deposit date: | 2004-02-02 | | Release date: | 2004-08-10 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and functional analysis of Mre11-3
Nucleic Acids Res., 32, 2004
|
|
2HIV
 
 | | ATP-dependent DNA ligase from S. solfataricus | | Descriptor: | Thermostable DNA ligase | | Authors: | Pascal, J.M, Ellenberger, T. | | Deposit date: | 2006-06-29 | | Release date: | 2006-11-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A Flexible Interface between DNA Ligase and PCNA Supports Conformational Switching and Efficient Ligation of DNA.
Mol.Cell, 24, 2006
|
|
2HIX
 
 | |
2FBV
 
 | | WRN exonuclease, Mn complex | | Descriptor: | MANGANESE (II) ION, Werner syndrome helicase | | Authors: | Perry, J.J. | | Deposit date: | 2005-12-10 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | WRN exonuclease structure and molecular mechanism imply an editing role in DNA end processing.
Nat.Struct.Mol.Biol., 13, 2006
|
|
2FBT
 
 | | WRN exonuclease | | Descriptor: | ACETIC ACID, Werner syndrome helicase | | Authors: | Perry, J.J. | | Deposit date: | 2005-12-10 | | Release date: | 2006-04-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | WRN exonuclease structure and molecular mechanism imply an editing role in DNA end processing.
Nat.Struct.Mol.Biol., 13, 2006
|
|
2G16
 
 | | Structure of S65A Y66S GFP variant after backbone fragmentation | | Descriptor: | Green fluorescent protein | | Authors: | Barondeau, D.P. | | Deposit date: | 2006-02-13 | | Release date: | 2006-04-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Understanding GFP Posttranslational Chemistry: Structures of Designed Variants that Achieve Backbone Fragmentation, Hydrolysis, and Decarboxylation.
J.Am.Chem.Soc., 128, 2006
|
|
2G6E
 
 | | Structure of cyclized F64L S65A Y66S GFP variant | | Descriptor: | Green fluorescent protein, MAGNESIUM ION | | Authors: | Barondeau, D.P. | | Deposit date: | 2006-02-24 | | Release date: | 2006-04-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Understanding GFP Posttranslational Chemistry: Structures of Designed Variants that Achieve Backbone Fragmentation, Hydrolysis, and Decarboxylation.
J.Am.Chem.Soc., 128, 2006
|
|
2G3D
 
 | |
2GDS
 
 | |
2G2S
 
 | |
2FBX
 
 | | WRN exonuclease, Mg complex | | Descriptor: | MAGNESIUM ION, Werner syndrome helicase | | Authors: | Perry, J.J. | | Deposit date: | 2005-12-10 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | WRN exonuclease structure and molecular mechanism imply an editing role in DNA end processing.
Nat.Struct.Mol.Biol., 13, 2006
|
|
2HIK
 
 | | heterotrimeric PCNA sliding clamp | | Descriptor: | PCNA1 (SSO0397), PCNA2 (SSO1047), PCNA3 (SSO0405) | | Authors: | Pascal, J.M, Tsodikov, O.V, Ellenberger, T. | | Deposit date: | 2006-06-29 | | Release date: | 2006-11-07 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | A Flexible Interface between DNA Ligase and PCNA Supports Conformational Switching and Efficient Ligation of DNA.
Mol.Cell, 24, 2006
|
|
2HII
 
 | | heterotrimeric PCNA sliding clamp | | Descriptor: | PCNA1 (SSO0397), PCNA2 (SSO1047), PCNA3 (SSO0405) | | Authors: | Pascal, J.M, Tsodikov, O.V, Ellenberger, T. | | Deposit date: | 2006-06-29 | | Release date: | 2006-11-07 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | A Flexible Interface between DNA Ligase and PCNA Supports Conformational Switching and Efficient Ligation of DNA.
Mol.Cell, 24, 2006
|
|
2JCW
 
 | | REDUCED BRIDGE-BROKEN YEAST CU/ZN SUPEROXIDE DISMUTASE ROOM TEMPERATURE (298K) STRUCTURE | | Descriptor: | COPPER (I) ION, CU/ZN SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Hart, P.J, Balbirnie, M.M, Ogihara, N.L, Nersissian, A.M, Weiss, M.S, Valentine, J.S, Eisenberg, D. | | Deposit date: | 1998-12-21 | | Release date: | 1999-06-08 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A structure-based mechanism for copper-zinc superoxide dismutase.
Biochemistry, 38, 1999
|
|
1F2T
 
 | | Crystal Structure of ATP-Free RAD50 ABC-ATPase | | Descriptor: | RAD50 ABC-ATPASE | | Authors: | Hopfner, K.P, Karcher, A, Shin, D.S, Craig, L. | | Deposit date: | 2000-05-29 | | Release date: | 2000-08-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily.
Cell(Cambridge,Mass.), 101, 2000
|
|
1F2U
 
 | | Crystal Structure of RAD50 ABC-ATPase | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, RAD50 ABC-ATPASE | | Authors: | Hopfner, K.P, Karcher, A, Shin, D.S, Craig, L. | | Deposit date: | 2000-05-29 | | Release date: | 2000-08-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily.
Cell(Cambridge,Mass.), 101, 2000
|
|
1SDA
 
 | | CRYSTAL STRUCTURE OF PEROXYNITRITE-MODIFIED BOVINE CU,ZN SUPEROXIDE DISMUTASE | | Descriptor: | COPPER (II) ION, COPPER,ZINC SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Smith, C.D, Carson, M, Van Der Woerd, M, Chen, J, Ischiropoulos, H, Beckman, J.S. | | Deposit date: | 1993-01-13 | | Release date: | 1993-10-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of peroxynitrite-modified bovine Cu,Zn superoxide dismutase.
Arch.Biochem.Biophys., 299, 1992
|
|
