9EC2
 
 | | Crystal structure of SAMHD1 dimer bound to an inhibitor obtained from high-throughput chemical tethering to the guanine antiviral acyclovir | | Descriptor: | Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, FE (III) ION, N-[5-({2-[(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)methoxy]ethyl}amino)-5-oxopentyl]-4,7-dibromo-3-hydroxynaphthalene-2-carboxamide | | Authors: | Egleston, M, Dong, L, Howlader, A.H, Bhat, S, Orris, B, Lopez-Rovira, L.M, Bianchet, M.A, Greenberg, M.M, Stivers, J.T. | | Deposit date: | 2024-11-13 | | Release date: | 2025-03-12 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Inhibitors of SAMHD1 Obtained from Chemical Tethering to the Guanine Antiviral Acyclovir.
Biochemistry, 64, 2025
|
|
9DYB
 
 | |
9DYA
 
 | | CHIP-TPR in complex with the Hsp70 tail | | Descriptor: | E3 ubiquitin-protein ligase CHIP, Heat shock 70 kDa protein 1A, SULFATE ION | | Authors: | Zhang, H, Nix, J.C, Page, R.C. | | Deposit date: | 2024-10-13 | | Release date: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Phosphorylation-State Modulated Binding of HSP70: Structural Insights and Compensatory Protein Engineering.
Biorxiv, 2025
|
|
9E6S
 
 | |
9EYS
 
 | | Structure of Far-Red Photosystem I from C. thermalis PCC 7203 | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Consoli, G, Tufaill, F, Murray, J.W, Fantuzzi, A, Rutherford, A.W. | | Deposit date: | 2024-04-09 | | Release date: | 2025-04-23 | | Method: | ELECTRON MICROSCOPY (2.01 Å) | | Cite: | Structure of Far-Red Photosystem I from C. thermalis PCC 7203
To Be Published
|
|
9E6T
 
 | |
9FTW
 
 | |
9F5U
 
 | | Crystal structure of Heme-Oxygenase from Corynebacterium diphtheriae complexed with Cobalt-porphyrine (HumO-Co(III)) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Labidi, R.J, Faivre, B, Carpentier, P, Perard, J, Gotico, P, Li, Y, Atta, M, Fontecave, M. | | Deposit date: | 2024-04-30 | | Release date: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Light-Activated Artificial CO 2 -Reductase: Structure and Activity.
J.Am.Chem.Soc., 2024
|
|
9F66
 
 | | Crystal structure of Heme-Oxygenase from Corynebacterium diphtheriae complexed with Cobalt-porphyrine (HumO-Co(III)) flash-cooled under CO2 pressure | | Descriptor: | 1,2-ETHANEDIOL, CARBON DIOXIDE, CHLORIDE ION, ... | | Authors: | Labidi, R.J, Faivre, B, Carpentier, P, Perard, J, Gotico, P, Li, Y, Atta, M, Fontecave, M. | | Deposit date: | 2024-04-30 | | Release date: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Light-Activated Artificial CO 2 -Reductase: Structure and Activity.
J.Am.Chem.Soc., 2024
|
|
9G3C
 
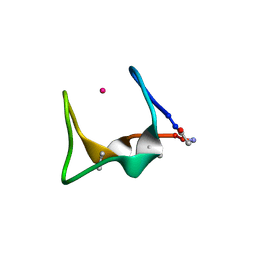 | |
9G39
 
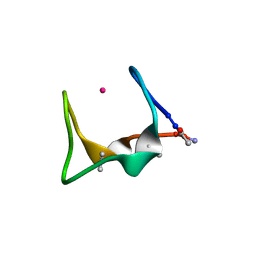 | |
9G3U
 
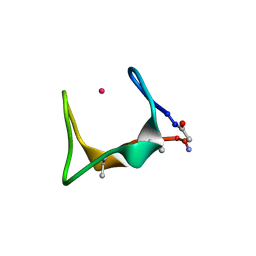 | |
9G3A
 
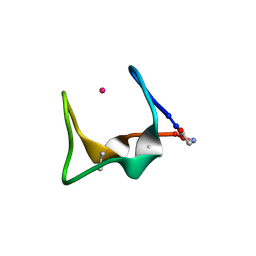 | | Crystal Structure of the artificial protein METP in complex with cadmium ion at different temperature, 160 K | | Descriptor: | CADMIUM ION, METP ARTIFICIAL PROTEIN | | Authors: | Di Costanzo, L, La Gatta, S, Chino, M. | | Deposit date: | 2024-07-12 | | Release date: | 2024-12-04 | | Last modified: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into temperature-dependent dynamics of METPsc1, a miniaturized electron-transfer protein.
J.Inorg.Biochem., 264, 2025
|
|
9GBF
 
 | | X-RAY structure of PHDvC5HCH tandem domain of NSD2 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, Histone-lysine N-methyltransferase NSD2, ... | | Authors: | Musco, G, Cocomazzi, P, Berardi, A, Knapp, S, Kramer, A. | | Deposit date: | 2024-07-31 | | Release date: | 2024-12-18 | | Last modified: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | The C-terminal PHDVC5HCH tandem domain of NSD2 is a combinatorial reader of unmodified H3K4 and tri-methylated H3K27 that regulates transcription of cell adhesion genes in multiple myeloma.
Nucleic Acids Res., 53, 2025
|
|
9FUO
 
 | | Crystal structure of SNAr1.3 (K39A) | | Descriptor: | CHLORIDE ION, Chain A, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Roberts, G.R, Leys, D. | | Deposit date: | 2024-06-26 | | Release date: | 2025-01-29 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Engineered enzymes for enantioselective nucleophilic aromatic substitutions.
Nature, 639, 2025
|
|
9FUL
 
 | |
9FCV
 
 | |
9F7O
 
 | |
9F7V
 
 | |
9G59
 
 | |
9FYV
 
 | |
9G65
 
 | |
9FYU
 
 | |
9FDH
 
 | | Closed Human phosphoglycerate kinase complex with BPG and ADP produced by cross-soaking a TSA crystal | | Descriptor: | 1,3-BISPHOSPHOGLYCERIC ACID, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Cliff, M.J, Waltho, J.P, Bowler, M.W, Baxter, N.J, Bisson, C, Blackburn, G.M. | | Deposit date: | 2024-05-17 | | Release date: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (1.756 Å) | | Cite: | The role of magnesium in catalysis by phosphoglycerate kinase
To be published
|
|
9FDN
 
 | | Human phosphoglycerate kinase in complex with ATP and 3PG formed by cross-soaking a TSA crystal | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, ADENOSINE-5'-TRIPHOSPHATE, Phosphoglycerate kinase 1, ... | | Authors: | Cliff, M.J, Waltho, J.P, Bowler, M.W, Baxter, N.J, Bisson, C, Blackburn, G.M. | | Deposit date: | 2024-05-17 | | Release date: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | The role of magnesium in catalysis by phosphoglycerate kinase
To be published
|
|
