2CV0
 
 | |
2CUZ
 
 | |
2CV2
 
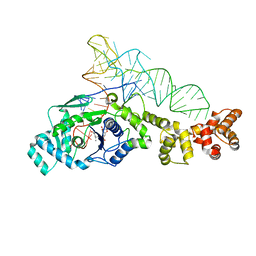 | | Glutamyl-tRNA synthetase from Thermus thermophilus in complex with tRNA(Glu) and an enzyme inhibitor, Glu-AMS | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, O5'-(L-GLUTAMYL-SULFAMOYL)-ADENOSINE, ... | | Authors: | Sekine, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-31 | | Release date: | 2006-09-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structural bases of transfer RNA-dependent amino acid recognition and activation by glutamyl-tRNA synthetase
Structure, 14, 2006
|
|
2DXI
 
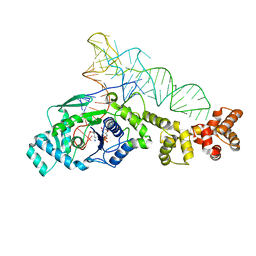 | | 2.2 A crystal structure of glutamyl-tRNA synthetase from Thermus thermophilus complexed with tRNA(Glu), ATP, and L-glutamol | | Descriptor: | (4S)-4-AMINO-5-HYDROXYPENTANOIC ACID, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Sekine, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-08-28 | | Release date: | 2006-10-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural bases of transfer RNA-dependent amino acid recognition and activation by glutamyl-tRNA synthetase
Structure, 14, 2006
|
|
6FTF
 
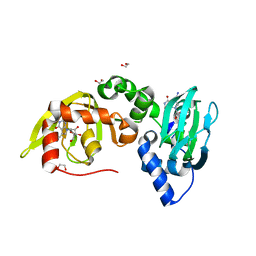 | | Regulatory subunit of a cAMP-independent protein kinase A from Trypanosoma cruzi at 1.09 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 7-cyano-7-deazainosine, Protein kinase A regulatory subunit, ... | | Authors: | Volpato Santos, Y, Lorentzen, E, Basquin, J, Boshart, M. | | Deposit date: | 2018-02-21 | | Release date: | 2019-04-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.09151936 Å) | | Cite: | Nucleoside analogue activators of cyclic AMP-independent protein kinase A of Trypanosoma.
Nat Commun, 10, 2019
|
|
8YJY
 
 | |
8WY4
 
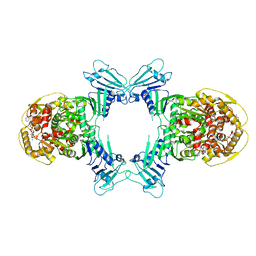 | | GajA tetramer with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Endonuclease GajA | | Authors: | Li, J, Wang, Z, Wang, L. | | Deposit date: | 2023-10-30 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Structures and activation mechanism of the Gabija anti-phage system.
Nature, 629, 2024
|
|
8X51
 
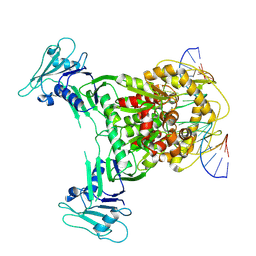 | | Structure of DNA-bound GajA dimer (focused refinement) | | Descriptor: | CALCIUM ION, DNA (5'-D(*AP*AP*AP*AP*AP*TP*AP*AP*CP*CP*GP*GP*GP*TP*TP*AP*TP*TP*AP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*AP*AP*TP*AP*AP*CP*CP*CP*GP*GP*TP*TP*AP*TP*TP*TP*TP*T)-3'), ... | | Authors: | Li, J, Wang, Z, Wang, L. | | Deposit date: | 2023-11-16 | | Release date: | 2024-02-28 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Structures and activation mechanism of the Gabija anti-phage system.
Nature, 629, 2024
|
|
8X5I
 
 | | Structure of ATP/Mg2+ bound Gabija GajA | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Endonuclease GajA, MAGNESIUM ION | | Authors: | Li, J, Wang, Z, Wang, L. | | Deposit date: | 2023-11-17 | | Release date: | 2024-02-28 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structures and activation mechanism of the Gabija anti-phage system.
Nature, 629, 2024
|
|
8X5N
 
 | | Structure of ATP/Mg2+ bound Gabija GajA-GajB 4:4 complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Endonuclease GajA, Gabija protein GajB, ... | | Authors: | Li, J, Wang, Z, Wang, L. | | Deposit date: | 2023-11-17 | | Release date: | 2024-02-28 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structures and activation mechanism of the Gabija anti-phage system.
Nature, 629, 2024
|
|
8WY5
 
 | | Structure of Gabija GajA in complex with DNA | | Descriptor: | CALCIUM ION, DNA (5'-D(P*AP*AP*AP*AP*TP*AP*AP*CP*CP*GP*GP*GP*TP*TP*AP*TP*TP*AP*A)-3'), DNA (5'-D(P*TP*TP*AP*AP*TP*AP*AP*CP*CP*CP*GP*GP*TP*TP*AP*TP*TP*TP*T)-3'), ... | | Authors: | Li, J, Wang, Z, Wang, L. | | Deposit date: | 2023-10-30 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Structures and activation mechanism of the Gabija anti-phage system.
Nature, 629, 2024
|
|
