3A3Y
 
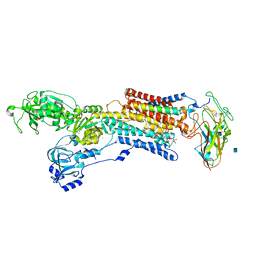 | | Crystal structure of the sodium-potassium pump with bound potassium and ouabain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, MAGNESIUM ION, ... | | Authors: | Ogawa, H, Shinoda, T, Cornelius, F, Toyoshima, C. | | Deposit date: | 2009-06-23 | | Release date: | 2009-09-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the sodium-potassium pump (Na+,K+-ATPase) with bound potassium and ouabain.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3A1E
 
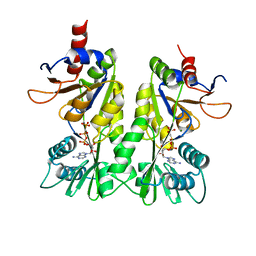 | | Crystal structure of the P- and N-domains of His462Gln mutant CopA, a copper-transporting P-type ATPase, bound with AMPPCP-Mg | | Descriptor: | MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Probable copper-exporting P-type ATPase A | | Authors: | Tsuda, T, Toyoshima, C. | | Deposit date: | 2009-03-31 | | Release date: | 2009-07-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Nucleotide recognition by CopA, a Cu+-transporting P-type ATPase.
Embo J., 28, 2009
|
|
3A1C
 
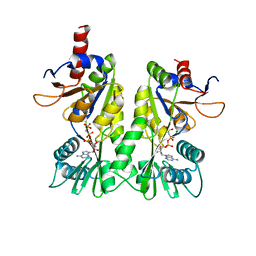 | | crystal structure of the P- and N-domains of CopA, a copper-transporting P-type ATPase, bound with AMPPCP-Mg | | Descriptor: | MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Probable copper-exporting P-type ATPase A | | Authors: | Tsuda, T, Toyoshima, C. | | Deposit date: | 2009-03-31 | | Release date: | 2009-07-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Nucleotide recognition by CopA, a Cu+-transporting P-type ATPase.
Embo J., 28, 2009
|
|
2EG9
 
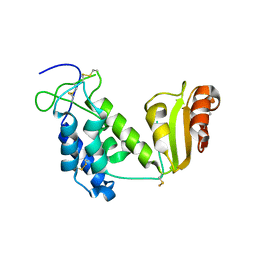 | | Crystal structure of the truncated extracellular domain of mouse CD38 | | Descriptor: | ADP-ribosyl cyclase 1 | | Authors: | Kukimoto-Niino, M, Mishima, C, Wakiyama, M, Terada, T, Shirouzu, M, Hara-Yokoyama, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-02-28 | | Release date: | 2008-03-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the truncated extracellular domain of mouse CD38
To be Published
|
|
3WGV
 
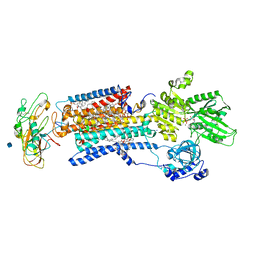 | | Crystal structure of a Na+-bound Na+,K+-ATPase preceding the E1P state with oligomycin | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Kanai, R, Ogawa, H, Vilsen, B, Cornelius, F, Toyoshima, C. | | Deposit date: | 2013-08-09 | | Release date: | 2013-10-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a Na1-bound Na1,K1-ATPase preceding the E1P state
Nature, 502, 2013
|
|
3WGU
 
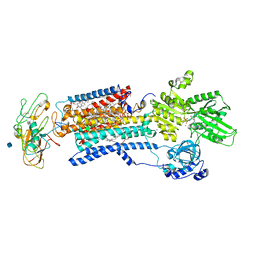 | | Crystal structure of a Na+-bound Na+,K+-ATPase preceding the E1P state without oligomycin | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Kanai, R, Ogawa, H, Vilsen, B, Cornelius, F, Toyoshima, C. | | Deposit date: | 2013-08-09 | | Release date: | 2013-10-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a Na1-bound Na1,K1-ATPase preceding the E1P state
Nature, 502, 2013
|
|
2D69
 
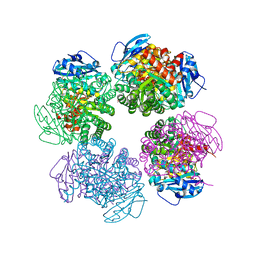 | | Crystal structure of the complex of sulfate ion and octameric ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) from Pyrococcus horikoshii OT3 (form-2 crystal) | | Descriptor: | Ribulose bisphosphate carboxylase, SULFATE ION | | Authors: | Mizohata, E, Mishima, C, Akasaka, R, Uda, H, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-11-10 | | Release date: | 2006-05-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the complex of sulfate ion and octameric ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) from Pyrococcus horikoshii OT3 (form-2 crystal)
To be Published
|
|
1WCY
 
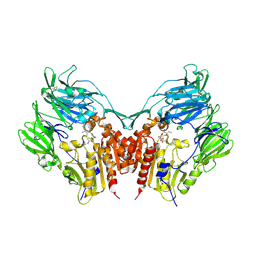 | | Crystal Structure Of Human Dipeptidyl Peptidase IV (DPPIV) Complex With Diprotin A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase IV, ... | | Authors: | Hiramatsu, H, Yamamoto, A, Kyono, K, Higashiyama, Y, Fukushima, C, Shima, H, Sugiyama, S, Inaka, K, Shimizu, R. | | Deposit date: | 2004-05-07 | | Release date: | 2005-05-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of human dipeptidyl peptidase IV (DPPIV) complex with diprotin A
Biol.Chem., 385, 2004
|
|
2EAT
 
 | | Crystal structure of the SR CA2+-ATPASE with bound CPA and TG | | Descriptor: | (6AR,11AS,11BR)-10-ACETYL-9-HYDROXY-7,7-DIMETHYL-2,6,6A,7,11A,11B-HEXAHYDRO-11H-PYRROLO[1',2':2,3]ISOINDOLO[4,5,6-CD]INDOL-11-ONE, OCTANOIC ACID [3S-[3ALPHA, 3ABETA, ... | | Authors: | Takahashi, M, Kondou, Y, Toyoshima, C. | | Deposit date: | 2007-02-02 | | Release date: | 2007-03-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Interdomain communication in calcium pump as revealed in the crystal structures with transmembrane inhibitors
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2EAU
 
 | | Crystal structure of the SR CA2+-ATPASE with bound CPA in the presence of curcumin | | Descriptor: | (6AR,11AS,11BR)-10-ACETYL-9-HYDROXY-7,7-DIMETHYL-2,6,6A,7,11A,11B-HEXAHYDRO-11H-PYRROLO[1',2':2,3]ISOINDOLO[4,5,6-CD]INDOL-11-ONE, PHOSPHATIDYLETHANOLAMINE, Sarcoplasmic/endoplasmic reticulum calcium ATPase 1 | | Authors: | Takahashi, M, Kondou, Y, Toyoshima, C. | | Deposit date: | 2007-02-02 | | Release date: | 2007-03-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Interdomain communication in calcium pump as revealed in the crystal structures with transmembrane inhibitors
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2EAR
 
 | | P21 crystal of the SR CA2+-ATPase with bound TG | | Descriptor: | OCTANOIC ACID [3S-[3ALPHA, 3ABETA, 4ALPHA, ... | | Authors: | Takahashi, M, Kondou, Y, Toyoshima, C. | | Deposit date: | 2007-02-02 | | Release date: | 2007-03-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Interdomain communication in calcium pump as revealed in the crystal structures with transmembrane inhibitors
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
1ULR
 
 | | Crystal structure of tt0497 from Thermus thermophilus HB8 | | Descriptor: | putative acylphosphatase | | Authors: | Ago, H, Hamada, K, Sugahara, M, Kuroishi, C, Kuramitsu, S, Yokoyama, S, Miyano, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-09-16 | | Release date: | 2004-11-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of tt0497 from Thermus thermophilus HB8
To be Published
|
|
1ULQ
 
 | | Crystal structure of tt0182 from Thermus thermophilus HB8 | | Descriptor: | putative acetyl-CoA acetyltransferase | | Authors: | Ago, H, Hamada, K, Ida, K, Kanda, H, Sugahara, M, Yamamoto, M, Kuroishi, C, Kuramitsu, S, Yokoyama, S, Miyano, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-09-16 | | Release date: | 2004-11-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of tt0182 from Thermus thermophilus HB8
To be Published
|
|
1YH3
 
 | | Crystal structure of human CD38 extracellular domain | | Descriptor: | ADP-ribosyl cyclase 1 | | Authors: | Liu, Q, Kriksunov, I.A, Graeff, R, Munshi, C, Lee, H.C, Hao, Q. | | Deposit date: | 2005-01-06 | | Release date: | 2005-09-27 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal structure of human CD38 extracellular domain.
Structure, 13, 2005
|
|
2DC0
 
 | | Crystal structure of amidase | | Descriptor: | probable amidase | | Authors: | Ohshima, T, Sakuraba, H, Ebihara, A, Kanagawa, M, Nakagawa, N, Kuroishi, C, Satoh, S, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-17 | | Release date: | 2007-01-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of amidase
To be Published
|
|
2E2Z
 
 | | Solution NMR structure of yeast Tim15, co-chaperone of mitochondrial Hsp70 | | Descriptor: | Tim15, ZINC ION | | Authors: | Momose, T, Ohshima, C, Maeda, M, Endo, T. | | Deposit date: | 2006-11-19 | | Release date: | 2007-10-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural basis of functional cooperation of Tim15/Zim17 with yeast mitochondrial Hsp70
Embo Rep., 8, 2007
|
|
2GOY
 
 | | Crystal structure of assimilatory adenosine 5'-phosphosulfate reductase with bound APS | | Descriptor: | ADENOSINE-5'-PHOSPHOSULFATE, IRON/SULFUR CLUSTER, adenosine phosphosulfate reductase | | Authors: | Chartron, J, Carroll, K.S, Shiau, C, Gao, H, Leary, J.A, Bertozzi, C.R, Stout, C.D. | | Deposit date: | 2006-04-14 | | Release date: | 2006-09-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Substrate Recognition, Protein Dynamics, and Iron-Sulfur Cluster in Pseudomonas aeruginosa Adenosine 5'-Phosphosulfate Reductase.
J.Mol.Biol., 364, 2006
|
|
2I67
 
 | | Structural Basis for the Mechanistic Understanding Human CD38 Controlled Multiple Catalysis | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, ADP-ribosyl cyclase 1 | | Authors: | Liu, Q, Kriksunov, I.A, Graeff, R, Munshi, C, Lee, H.C, Hao, Q. | | Deposit date: | 2006-08-28 | | Release date: | 2006-09-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structural basis for the mechanistic understanding of human CD38-controlled multiple catalysis.
J.Biol.Chem., 281, 2006
|
|
2I66
 
 | | Structural Basis for the Mechanistic Understanding Human CD38 Controlled Multiple Catalysis | | Descriptor: | ADP-ribosyl cyclase 1, [(2R,3R,4R,5R)-5-(2-AMINO-6-OXO-1,6-DIHYDRO-9H-PURIN-9-YL)-3,4-DIHYDROXYTETRAHYDROFURAN-2-YL]METHYL [(2R,3S,4R,5S)-3,4,5-TRIHYDROXYTETRAHYDROFURAN-2-YL]METHYL DIHYDROGEN DIPHOSPHATE, [(2R,3R,4R,5R)-5-(2-AMINO-6-OXO-1,6-DIHYDRO-9H-PURIN-9-YL)-3,4-DIHYDROXYTETRAHYDROFURAN-2-YL]METHYL [(2R,3S,4S)-3,4-DIHYDROXYTETRAHYDROFURAN-2-YL]METHYL DIHYDROGEN DIPHOSPHATE | | Authors: | Liu, Q, Kriksunov, I.A, Graeff, R, Munshi, C, Lee, H.C, Hao, Q. | | Deposit date: | 2006-08-28 | | Release date: | 2006-09-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for the mechanistic understanding of human CD38-controlled multiple catalysis.
J.Biol.Chem., 281, 2006
|
|
2I65
 
 | | Structural Basis for the Mechanistic Understanding Human CD38 Controlled Multiple Catalysis | | Descriptor: | ADP-ribosyl cyclase 1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Liu, Q, Kriksunov, I.A, Graeff, R, Munshi, C, Lee, H.C, Hao, Q. | | Deposit date: | 2006-08-28 | | Release date: | 2006-09-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the mechanistic understanding of human CD38-controlled multiple catalysis.
J.Biol.Chem., 281, 2006
|
|
3TFT
 
 | | Crystal structure of 7,8-diaminopelargonic acid synthase (BioA) from Mycobacterium tuberculosis, pre-reaction complex with a 3,6-dihydropyrid-2-one heterocycle inhibitor | | Descriptor: | Adenosylmethionine-8-amino-7-oxononanoate aminotransferase, DIMETHYL SULFOXIDE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Geders, T.W, Finzel, B.C. | | Deposit date: | 2011-08-16 | | Release date: | 2011-10-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Mechanism-based Inactivation by Aromatization of the Transaminase BioA Involved in Biotin Biosynthesis in Mycobaterium tuberculosis.
J.Am.Chem.Soc., 133, 2011
|
|
3TFU
 
 | | Crystal structure of 7,8-diaminopelargonic acid synthase (BioA) from Mycobacterium tuberculosis, post-reaction complex with a 3,6-dihydropyrid-2-one heterocycle inhibitor | | Descriptor: | Adenosylmethionine-8-amino-7-oxononanoate aminotransferase, DIMETHYL SULFOXIDE, [5-hydroxy-4-({[6-(3-hydroxypropyl)-2-oxo-1,2-dihydropyridin-3-yl]amino}methyl)-6-methylpyridin-3-yl]methyl dihydrogen phosphate | | Authors: | Geders, T.W, Finzel, B.C. | | Deposit date: | 2011-08-16 | | Release date: | 2011-10-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Mechanism-based Inactivation by Aromatization of the Transaminase BioA Involved in Biotin Biosynthesis in Mycobaterium tuberculosis.
J.Am.Chem.Soc., 133, 2011
|
|
6NK4
 
 | |
8T0N
 
 | | Structure of Compound 4 bound to human ALDH1A1 | | Descriptor: | 2-methoxy-6-{[(1-propyl-1H-benzimidazol-2-yl)amino]methyl}phenol, Aldehyde dehydrogenase 1A1, CHLORIDE ION, ... | | Authors: | Hurley, T.D. | | Deposit date: | 2023-06-01 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Development of substituted benzimidazoles as inhibitors of human aldehyde dehydrogenase 1A isoenzymes.
Chem.Biol.Interact., 391, 2024
|
|
8T0T
 
 | | Structure of Compound 4 bound to human ALDH1A1 | | Descriptor: | 1-(4-{6-fluoro-3-[4-(methanesulfonyl)piperazine-1-carbonyl]quinolin-4-yl}phenyl)cyclopropane-1-carbonitrile, Aldehyde dehydrogenase 1A1, CHLORIDE ION, ... | | Authors: | Hurley, T.D. | | Deposit date: | 2023-06-01 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Development of substituted benzimidazoles as inhibitors of human aldehyde dehydrogenase 1A isoenzymes.
Chem.Biol.Interact., 391, 2024
|
|
