8RA8
 
 | |
8R9G
 
 | |
8RA2
 
 | |
8R8J
 
 | |
8R9D
 
 | |
8R9Q
 
 | |
8RAE
 
 | |
8RAC
 
 | |
8R8X
 
 | |
4BGV
 
 | |
4BGU
 
 | | 1.50 A resolution structure of the malate dehydrogenase from Haloferax volcanii | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, CHLORIDE ION, ... | | Authors: | Talon, R, Madern, D, Girard, E. | | Deposit date: | 2013-03-28 | | Release date: | 2014-04-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.487 Å) | | Cite: | Insight Into Structural Evolution of Extremophilic Proteins
To be Published
|
|
4CL3
 
 | | 1.70 A resolution structure of the malate dehydrogenase from Chloroflexus aurantiacus | | Descriptor: | ACETATE ION, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Talon, R, Madern, D, Girard, E. | | Deposit date: | 2014-01-11 | | Release date: | 2014-02-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | An Experimental Point of View on Hydration/Solvation in Halophilic Proteins.
Front.Microbiol., 5, 2014
|
|
2V65
 
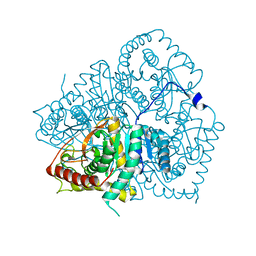 | | Apo LDH from the psychrophile C. gunnari | | Descriptor: | L-LACTATE DEHYDROGENASE A CHAIN | | Authors: | Coquelle, N, Madern, D, Vellieux, F. | | Deposit date: | 2007-07-13 | | Release date: | 2007-09-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Activity, Stability and Structural Studies of Lactate Dehydrogenases Adapted to Extreme Thermal Environments.
J.Mol.Biol., 374, 2007
|
|
2VK8
 
 | | Crystal structure of the Saccharomyces cerevisiae pyruvate decarboxylase variant E477Q in complex with its substrate | | Descriptor: | (2S)-2-HYDROXYPROPANOIC ACID, MAGNESIUM ION, PYRUVATE DECARBOXYLASE ISOZYME 1, ... | | Authors: | Kutter, S, Weik, M, Weiss, M.S, Konig, S. | | Deposit date: | 2007-12-17 | | Release date: | 2009-01-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Covalently Bound Substrate at the Regulatory Site of Yeast Pyruvate Decarboxylases Triggers Allosteric Enzyme Activation.
J.Biol.Chem., 284, 2009
|
|
2VK1
 
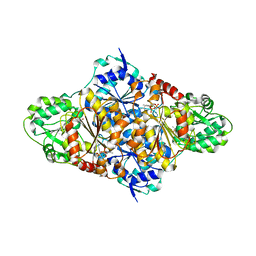 | | Crystal structure of the Saccharomyces cerevisiae pyruvate decarboxylase variant D28A in complex with its substrate | | Descriptor: | MAGNESIUM ION, PYRUVATE DECARBOXYLASE ISOZYME 1, PYRUVIC ACID, ... | | Authors: | Kutter, S, Weik, M, Weiss, M.S, Konig, S. | | Deposit date: | 2007-12-16 | | Release date: | 2009-01-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Covalently Bound Substrate at the Regulatory Site of Yeast Pyruvate Decarboxylases Triggers Allosteric Enzyme Activation.
J.Biol.Chem., 284, 2009
|
|
