8PIU
 
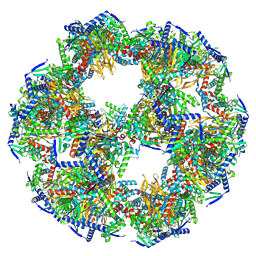 | | 60-meric complex of dihydrolipoamide acetyltransferase (E2) of the human pyruvate dehydrogenase complex | | Descriptor: | Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, mitochondrial | | Authors: | Zdanowicz, R, Afanasyev, P, Boehringer, D, Glockshuber, R. | | Deposit date: | 2023-06-22 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Stoichiometry and architecture of the human pyruvate dehydrogenase complex.
Sci Adv, 10, 2024
|
|
8RGX
 
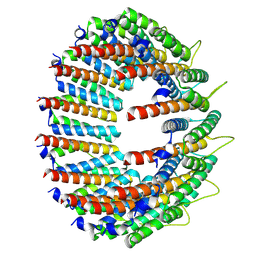 | | Cryo-EM structure of the Bacterial Proteasome Activator Bpa of Mycobacterium tuberculosis | | Descriptor: | Bacterial proteasome activator | | Authors: | Zdanowicz, R, von Rosen, T, Afanasyev, P, Boehringer, D, Glockshuber, R, Weber-Ban, E. | | Deposit date: | 2023-12-14 | | Release date: | 2025-04-02 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Substrates bind to residues lining the ring of asymmetrically engaged bacterial proteasome activator Bpa.
Nat Commun, 16, 2025
|
|
9FND
 
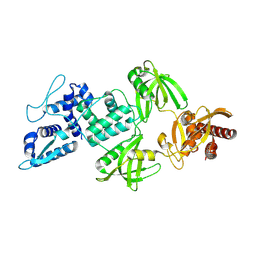 | | Transcriptional activator PafBC bound to mycobacterial RNA polymerase | | Descriptor: | PafC, Transcriptional regulator-like protein | | Authors: | Zdanowicz, R, Schilling, C.M, Rabl, J, Mueller, A.U, Boehringer, D, Glockshuber, R, Weber-Ban, E. | | Deposit date: | 2024-06-10 | | Release date: | 2025-02-12 | | Last modified: | 2025-02-19 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Single-stranded DNA binding to the transcription factor PafBC triggers the mycobacterial DNA damage response.
Sci Adv, 11, 2025
|
|
9FNE
 
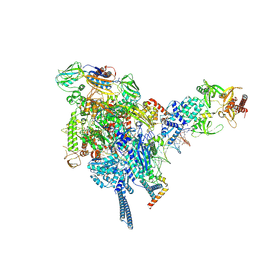 | | Mycobacterial PafBC-bound transcription initiation complex | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zdanowicz, R, Schilling, C.M, Rabl, J, Mueller, A.U, Boehringer, D, Glockshuber, R, Weber-Ban, E. | | Deposit date: | 2024-06-10 | | Release date: | 2025-02-12 | | Last modified: | 2025-02-19 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Single-stranded DNA binding to the transcription factor PafBC triggers the mycobacterial DNA damage response.
Sci Adv, 11, 2025
|
|
8ORB
 
 | |
8OQJ
 
 | |
8OSY
 
 | |
6Y7S
 
 | | 2.85 A cryo-EM structure of the in vivo assembled type 1 pilus rod | | Descriptor: | Type-1 fimbrial protein, A chain | | Authors: | Zyla, D, Hospenthal, M, Waksman, G, Glockshuber, R. | | Deposit date: | 2020-03-02 | | Release date: | 2021-03-31 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | The assembly platform FimD is required to obtain the most stable quaternary structure of type 1 pili.
Nat Commun, 15, 2024
|
|
8PSV
 
 | | 2.7 A cryo-EM structure of in vitro assembled type 1 pilus rod | | Descriptor: | Type-1 fimbrial protein, A chain | | Authors: | Hospenthal, M, Zyla, D, Glockshuber, R, Waksman, G. | | Deposit date: | 2023-07-13 | | Release date: | 2024-04-10 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | The assembly platform FimD is required to obtain the most stable quaternary structure of type 1 pili.
Nat Commun, 15, 2024
|
|
8PTU
 
 | | 2.5 A cryo-EM structure of the in vitro FimD-catalyzed assembly of type 1 pilus rod | | Descriptor: | Type-1 fimbrial protein, A chain | | Authors: | Zyla, D, Hospenthal, M, Glockshuber, R, Waksman, G. | | Deposit date: | 2023-07-14 | | Release date: | 2024-04-10 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | The assembly platform FimD is required to obtain the most stable quaternary structure of type 1 pili.
Nat Commun, 15, 2024
|
|
