4B56
 
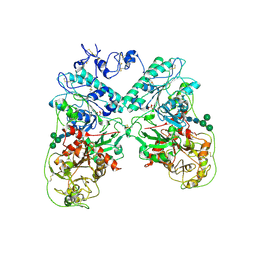 | | Structure of ectonucleotide pyrophosphatase-phosphodiesterase-1 (NPP1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Jansen, S, Perrakis, A, Ulens, C, Winkler, C, Andries, M, Joosten, R.P, Van Acker, M, Luyten, F.P, Moolenaar, W.H, Bollen, M. | | Deposit date: | 2012-08-02 | | Release date: | 2012-09-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of Npp1, an Ectonucleotide Pyrophosphatase/Phosphodiesterase Involved in Tissue Calcification.
Structure, 20, 2012
|
|
3ZOH
 
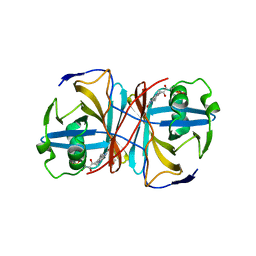 | | Crystal structure of FMN-binding protein (YP_005476) from Thermus thermophilus with bound 1-Cyclohex-2-enone | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVOREDOXIN, cyclohex-2-en-1-one | | Authors: | Pavkov-Keller, T, Steinkellner, G, Gruber, C.C, Steiner, K, Winkler, C, Schwamberger, O, Schwab, H, Faber, K, Gruber, K. | | Deposit date: | 2013-02-21 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Identification of Promiscuous Ene-Reductase Activity by Mining Structural Databases Using Active Site Constellations.
Nat.Commun., 5, 2014
|
|
3ZOC
 
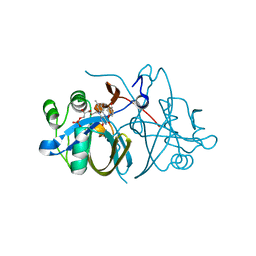 | | Crystal structure of FMN-binding protein (NP_142786.1) from Pyrococcus horikoshii with bound p-hydroxybenzaldehyde | | Descriptor: | FLAVIN MONONUCLEOTIDE, FMN-BINDING PROTEIN, P-HYDROXYBENZALDEHYDE | | Authors: | Pavkov-Keller, T, Steinkellner, G, Gruber, C.C, Steiner, K, Winkler, C, Schwamberger, O, Schwab, H, Faber, K, Gruber, K. | | Deposit date: | 2013-02-21 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of Promiscuous Ene-Reductase Activity by Mining Structural Databases Using Active Site Constellations.
Nat.Commun., 5, 2014
|
|
3ZOE
 
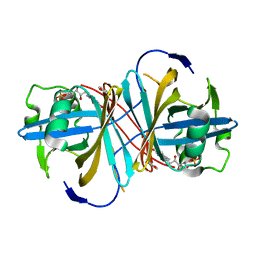 | | Crystal structure of FMN-binding protein (YP_005476) from Thermus thermophilus with bound p-hydroxybenzaldehyde | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVOREDOXIN, P-HYDROXYBENZALDEHYDE | | Authors: | Pavkov-Keller, T, Steinkellner, G, Gruber, C.C, Steiner, K, Winkler, C, Schwamberger, O, Schwab, H, Faber, K, Gruber, K. | | Deposit date: | 2013-02-21 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Identification of Promiscuous Ene-Reductase Activity by Mining Structural Databases Using Active Site Constellations.
Nat.Commun., 5, 2014
|
|
3ZOD
 
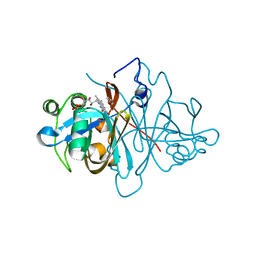 | | Crystal structure of FMN-binding protein (NP_142786.1) from Pyrococcus horikoshii with bound benzene-1,4-diol | | Descriptor: | FLAVIN MONONUCLEOTIDE, FMN-BINDING PROTEIN, benzene-1,4-diol | | Authors: | Pavkov-Keller, T, Steinkellner, G, Gruber, C.C, Steiner, K, Winkler, C, Schwamberger, O, Schwab, H, Faber, K, Gruber, K. | | Deposit date: | 2013-02-21 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Identification of Promiscuous Ene-Reductase Activity by Mining Structural Databases Using Active Site Constellations.
Nat.Commun., 5, 2014
|
|
3ZOG
 
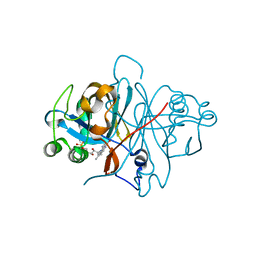 | | Crystal structure of FMN-binding protein (NP_142786.1) from Pyrococcus horikoshii with bound 1-Cyclohex-2-enone | | Descriptor: | FLAVIN MONONUCLEOTIDE, FMN-BINDING PROTEIN, cyclohex-2-en-1-one | | Authors: | Pavkov-Keller, T, Steinkellner, G, Gruber, C.C, Steiner, K, Winkler, C, Schwamberger, O, Schwab, H, Faber, K, Gruber, K. | | Deposit date: | 2013-02-21 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Identification of Promiscuous Ene-Reductase Activity by Mining Structural Databases Using Active Site Constellations.
Nat.Commun., 5, 2014
|
|
3ZOF
 
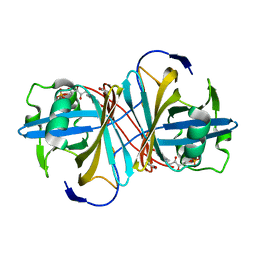 | | Crystal structure of FMN-binding protein (YP_005476) from Thermus thermophilus with bound benzene-1,4-diol | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVOREDOXIN, benzene-1,4-diol | | Authors: | Pavkov-Keller, T, Steinkellner, G, Gruber, C.C, Steiner, K, Winkler, C, Schwamberger, O, Schwab, H, Faber, K, Gruber, K. | | Deposit date: | 2013-02-21 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Identification of Promiscuous Ene-Reductase Activity by Mining Structural Databases Using Active Site Constellations.
Nat.Commun., 5, 2014
|
|
5JVJ
 
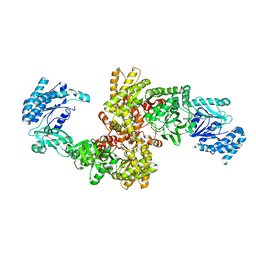 | | C4-type pyruvate phosphate dikinase: different conformational states of the nucleotide binding domain in the dimer | | Descriptor: | MAGNESIUM ION, PHOSPHOENOLPYRUVATE, Pyruvate, ... | | Authors: | Minges, A, Hoeppner, A, Groth, G. | | Deposit date: | 2016-05-11 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.898 Å) | | Cite: | Structural intermediates and directionality of the swiveling motion of Pyruvate Phosphate Dikinase.
Sci Rep, 7, 2017
|
|
5JVN
 
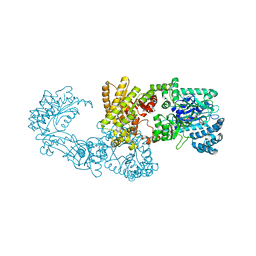 | | C3-type pyruvate phosphate dikinase: intermediate state of the central domain in the swiveling mechanism | | Descriptor: | 2'-Bromo-2'-deoxyadenosine 5'-[beta,gamma-imide]triphosphoric acid, MAGNESIUM ION, PHOSPHOENOLPYRUVATE, ... | | Authors: | Minges, A, Hoeppner, A, Groth, G. | | Deposit date: | 2016-05-11 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural intermediates and directionality of the swiveling motion of Pyruvate Phosphate Dikinase.
Sci Rep, 7, 2017
|
|
5JVL
 
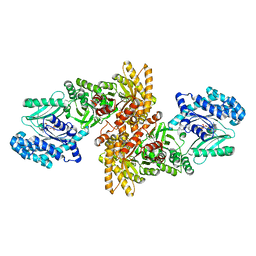 | | C4-type pyruvate phospate dikinase: nucleotide binding domain with bound ATP analogue | | Descriptor: | 2'-Bromo-2'-deoxyadenosine 5'-[beta,gamma-imide]triphosphoric acid, MAGNESIUM ION, PHOSPHOENOLPYRUVATE, ... | | Authors: | Minges, A, Hoeppner, A, Groth, G. | | Deposit date: | 2016-05-11 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural intermediates and directionality of the swiveling motion of Pyruvate Phosphate Dikinase.
Sci Rep, 7, 2017
|
|
2LUT
 
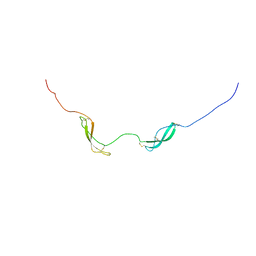 | | NMR solution structure of midkine-a | | Descriptor: | Midkine-related growth factor | | Authors: | Lim, J, Yang, D, Meng, D. | | Deposit date: | 2012-06-21 | | Release date: | 2013-05-01 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Structural and Functional Characterization of Two Zebrafish Midkine Proteins Reveals Importance of the Conserved Hinge for Heparin Binding and Embryogenesis
To be Published
|
|
2LUU
 
 | | NMR solution structure of midkine-b, mdkb | | Descriptor: | Midkine-related growth factor Mdk2 | | Authors: | Yang, D, Lim, J, Meng, D. | | Deposit date: | 2012-06-21 | | Release date: | 2013-05-01 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Structure-function analysis of full-length midkine reveals novel residues important for heparin binding and zebrafish embryogenesis.
Biochem.J., 451, 2013
|
|
