3VUA
 
 | | Apo IsdH-NEAT3 in space group P3121 at a resolution of 1.85 A | | Descriptor: | ACETATE ION, GLYCEROL, Iron-regulated surface determinant protein H, ... | | Authors: | Vu, N.T, Caaveiro, J.M.M, Moriwaki, Y, Tsumoto, K. | | Deposit date: | 2012-06-26 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of heme transport protein IsdH-NEAT3 from S. aureus in complex with Indium-porphyrin
To be Published
|
|
3VTM
 
 | | Structure of heme transport protein IsdH-NEAT3 from S. aureus in complex with Indium-porphyrin | | Descriptor: | GLYCEROL, Iron-regulated surface determinant protein H, PROTOPORPHYRIN IX CONTAINING INDIUM | | Authors: | Vu, N.T, Caaveiro, J.M.M, Moriwaki, Y, Tsumoto, K. | | Deposit date: | 2012-05-31 | | Release date: | 2013-05-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Selective binding of antimicrobial porphyrins to the heme-receptor IsdH-NEAT3 of Staphylococcus aureus
Protein Sci., 22, 2013
|
|
6IEJ
 
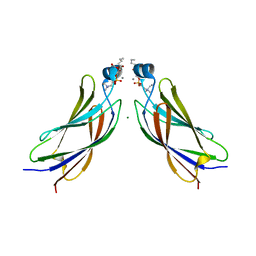 | | The C2 domain of cytosolic phospholipase A2 alpha bound to phosphatidylcholine | | Descriptor: | 1,2-dihexanoyl-sn-glycero-3-phosphocholine, CALCIUM ION, Cytosolic phospholipase A2, ... | | Authors: | Hirano, Y, Gao, Y.G, Stephenson, D.J, Vu, N.T, Malinina, L, Chalfant, C.E, Patel, D.J, Brown, R.E. | | Deposit date: | 2018-09-14 | | Release date: | 2019-05-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.206 Å) | | Cite: | Structural basis of phosphatidylcholine recognition by the C2-domain of cytosolic phospholipase A2alpha.
Elife, 8, 2019
|
|
3AMK
 
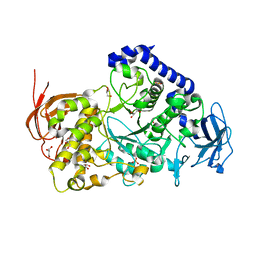 | | Structure of the Starch Branching Enzyme I (BEI) from Oryza sativa L | | Descriptor: | GLYCEROL, Os06g0726400 protein, PHOSPHATE ION | | Authors: | Kakuta, Y, Chaen, K, Noguchi, J, Vu, N, Kimura, M. | | Deposit date: | 2010-08-20 | | Release date: | 2011-09-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the branching enzyme I (BEI) from Oryza sativa L with implications for catalysis and substrate binding.
Glycobiology, 21, 2011
|
|
3AML
 
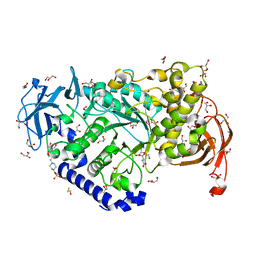 | | Structure of the Starch Branching Enzyme I (BEI) from Oryza sativa L | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, BETA-MERCAPTOETHANOL, ... | | Authors: | Kakuta, Y, Chaen, K, Noguchi, J, Vu, N, Kimura, M. | | Deposit date: | 2010-08-20 | | Release date: | 2011-09-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the branching enzyme I (BEI) from Oryza sativa L with implications for catalysis and substrate binding.
Glycobiology, 21, 2011
|
|
