5XZE
 
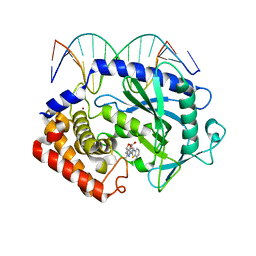 | | Mouse cGAS bound to the inhibitor RU332 | | Descriptor: | (3R)-3-[1-(3H-1lambda~4~,3-benzothiazol-2-yl)-5-hydroxy-3-methyl-1H-pyrazol-4-yl]-2-benzofuran-1(3H)-one, Cyclic GMP-AMP synthase, DNA (5'-D(*AP*AP*AP*TP*TP*GP*CP*CP*GP*AP*AP*GP*AP*CP*G)-3'), ... | | Authors: | Vincent, J, Adura, C, Gao, P, Luz, A, Lama, L, Asano, Y, Okamoto, R, Imaeda, T, Aida, J, Rothamel, K, Gogakos, T, Steinberg, J, Reasoner, S, Aso, K, Tuschl, T, Patel, D.J, Glickman, J.F, Ascano, M. | | Deposit date: | 2017-07-12 | | Release date: | 2017-10-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.177 Å) | | Cite: | Small molecule inhibition of cGAS reduces interferon expression in primary macrophages from autoimmune mice.
Nat Commun, 8, 2017
|
|
5XZG
 
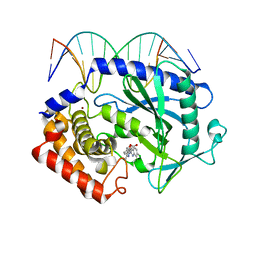 | | Mouse cGAS bound to the inhibitor RU521 | | Descriptor: | 2-(4,5-dichloro-1H-benzimidazol-2-yl)-5-methyl-4-[(1R)-3-oxo-1,3-dihydro-2-benzofuran-1-yl]-1,2-dihydro-3H-pyrazol-3-one, Cyclic GMP-AMP synthase, DNA (5'-D(*AP*AP*AP*TP*TP*GP*CP*CP*GP*AP*AP*GP*AP*CP*G)-3'), ... | | Authors: | Vincent, J, Adura, C, Gao, P, Luz, A, Lama, L, Asano, Y, Okamoto, R, Imaeda, T, Aida, J, Rothamel, K, Gogakos, T, Steinberg, J, Reasoner, S, Aso, K, Tuschl, T, Patel, D.J, Glickman, J.F, Ascano, M. | | Deposit date: | 2017-07-12 | | Release date: | 2017-10-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.828 Å) | | Cite: | Small molecule inhibition of cGAS reduces interferon expression in primary macrophages from autoimmune mice.
Nat Commun, 8, 2017
|
|
5XZB
 
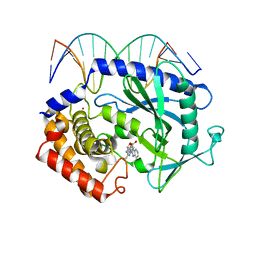 | | Mouse cGAS bound to the inhibitor RU365 | | Descriptor: | (3R)-3-[1-(1H-benzimidazol-2-yl)-5-hydroxy-3-methyl-1H-pyrazol-4-yl]-2-benzofuran-1(3H)-one, Cyclic GMP-AMP synthase, DNA (5'-D(*AP*AP*AP*TP*TP*GP*CP*CP*GP*AP*AP*GP*AP*CP*G)-3'), ... | | Authors: | Vincent, J, Adura, C, Gao, P, Luz, A, Lama, L, Asano, Y, Okamoto, R, Imaeda, T, Aida, J, Rothamel, K, Gogakos, T, Steinberg, J, Reasoner, S, Aso, K, Tuschl, T, Patel, D.J, Glickman, J.F, Ascano, M. | | Deposit date: | 2017-07-12 | | Release date: | 2017-08-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Small molecule inhibition of cGAS reduces interferon expression in primary macrophages from autoimmune mice.
Nat Commun, 8, 2017
|
|
1S96
 
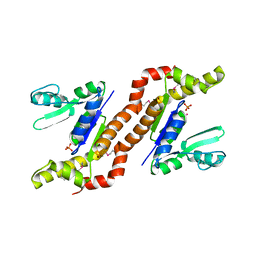 | | The 2.0 A X-ray structure of Guanylate Kinase from E.coli | | Descriptor: | Guanylate kinase, PHOSPHATE ION, UNKNOWN ATOM OR ION | | Authors: | Kreusch, A, Spraggon, G, Klock, H.E, McMullan, D, Vincent, J, Rodrigues, K, Lesley, S.A. | | Deposit date: | 2004-02-03 | | Release date: | 2004-02-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Structure of Guanylate Kinase from Escherichia coli at 2.0 A resolution
To be Published
|
|
2X5V
 
 | | 80 microsecond laue diffraction snapshot from crystals of a photosynthetic reaction centre 3 millisecond following photoactivation. | | Descriptor: | BACTERIOCHLOROPHYLL B, BACTERIOPHEOPHYTIN B, FE (II) ION, ... | | Authors: | Wohri, A.B, Katona, G, Johansson, L.C, Fritz, E, Malmerberg, E, Andersson, M, Vincent, J, Eklund, M, Cammarata, M, Wulff, M, Davidsson, J, Groenhof, G, Neutze, R. | | Deposit date: | 2010-02-10 | | Release date: | 2010-05-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Light-Induced Structural Changes in a Photosynthetic Reaction Center Caught by Laue Diffraction.
Science, 328, 2010
|
|
2X5U
 
 | | 80 microsecond Laue diffraction snapshot from crystals of a photosynthetic reaction centre without illumination. | | Descriptor: | BACTERIOCHLOROPHYLL B, BACTERIOPHEOPHYTIN B, FE (II) ION, ... | | Authors: | Wohri, A.B, Katona, G, Johansson, L.C, Fritz, E, Malmerberg, E, Andersson, M, Vincent, J, Eklund, M, Cammarata, M, Wulff, M, Davidsson, J, Groenhof, G, Neutze, R. | | Deposit date: | 2010-02-10 | | Release date: | 2010-05-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Light-induced structural changes in a photosynthetic reaction center caught by Laue diffraction.
Science, 328, 2010
|
|
1LM4
 
 | | Structure of Peptide Deformylase from Staphylococcus aureus at 1.45 A | | Descriptor: | FE (III) ION, GLYCEROL, peptide deformylase PDF1 | | Authors: | Kreusch, A, Spraggon, G, Lee, C.C, Klock, H, McMullan, D, Ng, K, Shin, T, Vincent, J, Warner, I, Ericson, C, Lesley, S.A. | | Deposit date: | 2002-04-30 | | Release date: | 2003-06-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure analysis of peptide deformylases from streptococcus pneumoniae,staphylococcus aureus, thermotoga maritima, and pseudomonas aeruginosa: snapshots of the oxygen sensitivity of peptide deformylase
J.MOL.BIOL., 330, 2003
|
|
1LME
 
 | | Crystal Structure of Peptide Deformylase from Thermotoga maritima | | Descriptor: | peptide deformylase | | Authors: | Kreusch, A, Spraggon, G, Lee, C.C, Klock, H, McMullan, D, Ng, K, Shin, T, Vincent, J, Warner, I, Ericson, C, Lesley, S.A, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2002-05-01 | | Release date: | 2003-06-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure analysis of peptide deformylases from streptococcus pneumoniae,staphylococcus aureus, thermotoga maritima, and pseudomonas aeruginosa: snapshots of the oxygen sensitivity of peptide deformylase
J.MOL.BIOL., 330, 2003
|
|
1LM6
 
 | | Crystal Structure of Peptide Deformylase from Streptococcus pneumoniae | | Descriptor: | FE (III) ION, GLYCEROL, peptide deformylase DEFB | | Authors: | Kreusch, A, Spraggon, G, Lee, C.C, Klock, H, McMullan, D, Ng, K, Shin, T, Vincent, J, Warner, I, Ericson, C, Lesley, S.A. | | Deposit date: | 2002-04-30 | | Release date: | 2003-06-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure analysis of peptide deformylases from streptococcus pneumoniae,staphylococcus aureus, thermotoga maritima, and pseudomonas aeruginosa: snapshots of the oxygen sensitivity of peptide deformylase
J.MOL.BIOL., 330, 2003
|
|
1N5N
 
 | | Crystal Structure of Peptide Deformylase from Pseudomonas aeruginosa | | Descriptor: | GLYCEROL, Peptide deformylase, ZINC ION | | Authors: | Kreusch, A, Spraggon, G, Lee, C.C, Klock, H, McMullan, D, Ng, K, Shin, T, Vincent, J, Warner, I, Ericson, C, Lesley, S.A. | | Deposit date: | 2002-11-06 | | Release date: | 2003-06-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure analysis of peptide deformylases from streptococcus pneumoniae,staphylococcus aureus, thermotoga maritima, and pseudomonas aeruginosa: snapshots of the oxygen sensitivity of peptide deformylase
J.MOL.BIOL., 330, 2003
|
|
1J6U
 
 | |
4UZ9
 
 | | STRUCTURE OF THE WNT DEACYLASE NOTUM - CRYSTAL FORM VII - SOS COMPLEX - 2.2A | | Descriptor: | 1,3,4,6-tetra-O-sulfo-beta-D-fructofuranose-(2-1)-2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Zebisch, M, Jones, E.Y. | | Deposit date: | 2014-09-04 | | Release date: | 2015-02-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Notum Deacylates Wnt Proteins to Suppress Signalling Activity.
Nature, 519, 2015
|
|
4UZL
 
 | |
4UYW
 
 | |
1S17
 
 | | Identification of Novel Potent Bicyclic Peptide Deformylase Inhibitors | | Descriptor: | 2-(3,4-DIHYDRO-3-OXO-2H-BENZO[B][1,4]THIAZIN-2-YL)-N-HYDROXYACETAMIDE, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Molteni, V, He, X, Nabakka, J, Yang, K, Kreusch, A, Gordon, P, Bursulaya, B, Ryder, N.S, Goldberg, R, He, Y. | | Deposit date: | 2004-01-05 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Identification of novel potent bicyclic peptide deformylase inhibitors
Bioorg.Med.Chem.Lett., 14, 2004
|
|
1XNX
 
 | | Crystal structure of constitutive androstane receptor | | Descriptor: | 16,17-ANDROSTENE-3-OL, constitutive androstane receptor | | Authors: | Fernandez, E. | | Deposit date: | 2004-10-05 | | Release date: | 2005-01-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the murine constitutive androstane receptor complexed to androstenol; a molecular basis for inverse agonism
Mol.Cell, 16, 2004
|
|
4UZ1
 
 | |
4UYZ
 
 | |
4UZA
 
 | |
4UZK
 
 | |
4UZJ
 
 | |
4UYU
 
 | |
4UZ5
 
 | |
4UZ6
 
 | | STRUCTURE OF THE WNT DEACYLASE NOTUM - CRYSTAL FORM V - SOS COMPLEX - 1.9A | | Descriptor: | 1,3,4,6-tetra-O-sulfo-beta-D-fructofuranose-(2-1)-2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Zebisch, M, Jones, E.Y. | | Deposit date: | 2014-09-04 | | Release date: | 2015-02-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Notum Deacylates Wnt Proteins to Suppress Signalling Activity.
Nature, 519, 2015
|
|
4UZQ
 
 | |
