3CB6
 
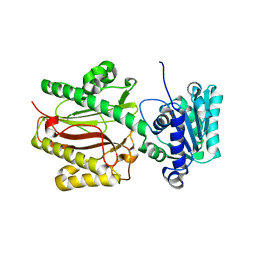 | | Crystal Structure of the S. pombe Peptidase Homology Domain of FACT complex subunit Spt16 (form B) | | Descriptor: | FACT complex subunit spt16 | | Authors: | Stuwe, T, Hothorn, M, Lejeune, E, Bortfeld-Miller, M, Scheffzek, K, Ladurner, A.G. | | Deposit date: | 2008-02-21 | | Release date: | 2008-06-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | The FACT Spt16 "peptidase" domain is a histone H3-H4 binding module
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3CB5
 
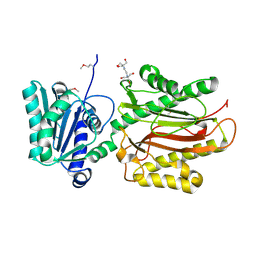 | | Crystal Structure of the S. pombe Peptidase Homology Domain of FACT complex subunit Spt16 (form A) | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FACT complex subunit spt16 | | Authors: | Stuwe, T, Hothorn, M, Lejeune, E, Bortfeld-Miller, M, Scheffzek, K, Ladurner, A.G. | | Deposit date: | 2008-02-21 | | Release date: | 2008-06-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The FACT Spt16 "peptidase" domain is a histone H3-H4 binding module
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
4XMN
 
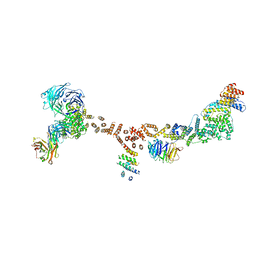 | | Structure of the yeast coat nucleoporin complex, space group P212121 | | Descriptor: | Antibody 87 heavy chain, Antibody 87 light chain, Nucleoporin NUP120, ... | | Authors: | Stuwe, T, Correia, A.R, Lin, D.H, Paduch, M, Lu, V.T, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2015-01-14 | | Release date: | 2015-03-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (7.6 Å) | | Cite: | Nuclear pores. Architecture of the nuclear pore complex coat.
Science, 347, 2015
|
|
4XMM
 
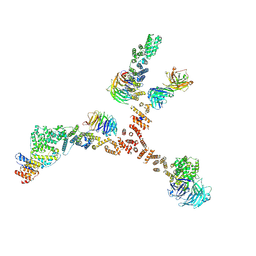 | | Structure of the yeast coat nucleoporin complex, space group C2 | | Descriptor: | Antibody 57 heavy chain, Antibody 57 light chain, Nucleoporin NUP120, ... | | Authors: | Stuwe, T, Correia, A.R, Lin, D.H, Paduch, M, Lu, V.T, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2015-01-14 | | Release date: | 2015-03-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (7.384 Å) | | Cite: | Nuclear pores. Architecture of the nuclear pore complex coat.
Science, 347, 2015
|
|
5CWV
 
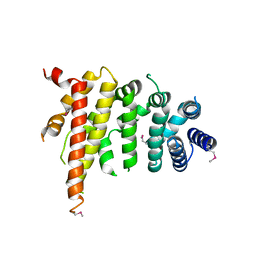 | | Crystal structure of Chaetomium thermophilum Nup192 TAIL domain | | Descriptor: | Nucleoporin NUP192 | | Authors: | Stuwe, T, Bley, C.J, Thierbach, K, Petrovic, S, Schilbach, S, Mayo, D.J, Perriches, T, Rundlet, E.J, Jeon, Y.E, Collins, L.N, Lin, D.H, Paduch, M, Koide, A, Lu, V, Fischer, J, Hurt, E, Koide, S, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2015-07-28 | | Release date: | 2015-09-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.155 Å) | | Cite: | Architecture of the fungal nuclear pore inner ring complex.
Science, 350, 2015
|
|
5CWU
 
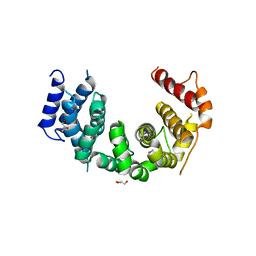 | | Crystal structure of Chaetomium thermophilum Nup188 TAIL domain | | Descriptor: | GLYCEROL, Nucleoporin NUP188 | | Authors: | Stuwe, T, Bley, C.J, Thierbach, K, Petrovic, S, Schilbach, S, Mayo, D.J, Perriches, T, Rundlet, E.J, Jeon, Y.E, Collins, L.N, Lin, D.H, Paduch, M, Koide, A, Lu, V, Fischer, J, Hurt, E, Koide, S, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2015-07-28 | | Release date: | 2015-09-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Architecture of the fungal nuclear pore inner ring complex.
Science, 350, 2015
|
|
5CWW
 
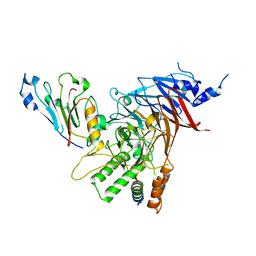 | | Crystal structure of the Chaetomium thermophilum heterotrimeric Nup82 NTD-Nup159 TAIL-Nup145N APD complex | | Descriptor: | Nucleoporin NUP145N, Nucleoporin NUP159, Nucleoporin NUP82 | | Authors: | Stuwe, T, Bley, C.J, Thierbach, K, Petrovic, S, Schilbach, S, Mayo, D.J, Perriches, T, Rundlet, E.J, Jeon, Y.E, Collins, L.N, Lin, D.H, Paduch, M, Koide, A, Lu, V, Fischer, J, Hurt, E, Koide, S, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2015-07-28 | | Release date: | 2015-09-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Architecture of the fungal nuclear pore inner ring complex.
Science, 350, 2015
|
|
4JQ5
 
 | |
4JNU
 
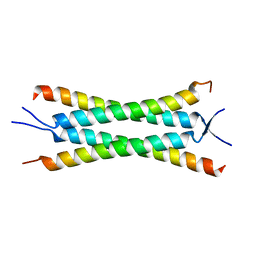 | | Crystal structure of the human Nup57CCS3* coiled-coil segment, space group P21 | | Descriptor: | Nucleoporin p54 | | Authors: | Stuwe, T, Bley, C.J, Mayo, D.J, Hoelz, A. | | Deposit date: | 2013-03-15 | | Release date: | 2014-09-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.445 Å) | | Cite: | Architecture of the fungal nuclear pore inner ring complex.
Science, 350, 2015
|
|
4JNV
 
 | | Crystal structure of the human Nup57CCS3* coiled-coil segment, space group C2 | | Descriptor: | Nucleoporin p54 | | Authors: | Stuwe, T, Bley, C.J, Mayo, D.J, Hoelz, A. | | Deposit date: | 2013-03-15 | | Release date: | 2014-09-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Architecture of the fungal nuclear pore inner ring complex.
Science, 350, 2015
|
|
4JO7
 
 | | Crystal structure of the human Nup49CCS2+3* Nup57CCS3* complex with 2:2 stoichiometry | | Descriptor: | Nucleoporin p54, Nucleoporin p58/p45 | | Authors: | Stuwe, T, Bley, C.J, Mayo, D.J, Hoelz, A. | | Deposit date: | 2013-03-17 | | Release date: | 2014-09-17 | | Last modified: | 2016-02-03 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Architecture of the fungal nuclear pore inner ring complex.
Science, 350, 2015
|
|
4JO9
 
 | | Crystal structure of the human Nup49CCS2+3* Nup57CCS3* complex 1:2 stoichiometry | | Descriptor: | Nucleoporin p54, Nucleoporin p58/p45 | | Authors: | Stuwe, T, Bley, C.J, Mayo, D.J, Hoelz, A. | | Deposit date: | 2013-03-18 | | Release date: | 2014-09-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | Architecture of the fungal nuclear pore inner ring complex.
Science, 350, 2015
|
|
4KNH
 
 | | Structure of the Chaetomium thermophilum adaptor nucleoporin Nup192 N-terminal domain | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Nup192p, ... | | Authors: | Stuwe, T, Lin, D.H, Collins, L.N, Hoelz, A. | | Deposit date: | 2013-05-09 | | Release date: | 2014-02-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.702 Å) | | Cite: | Evidence for an evolutionary relationship between the large adaptor nucleoporin Nup192 and karyopherins.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4KHB
 
 | | Structure of the Spt16D Pob3N heterodimer | | Descriptor: | SULFATE ION, Uncharacterized protein POB3N, Uncharacterized protein SPT16D | | Authors: | Stuwe, T, Zhang, E, Ladurner, A.G. | | Deposit date: | 2013-04-30 | | Release date: | 2013-05-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of histone H2A-H2B recognition by the essential chaperone FACT.
Nature, 499, 2013
|
|
4KHO
 
 | |
5HB4
 
 | |
4I9Y
 
 | | Structure of the C-terminal domain of Nup358 | | Descriptor: | CHLORIDE ION, E3 SUMO-protein ligase RanBP2, GLYCEROL, ... | | Authors: | Lin, D.H, Zimmermann, S, Stuwe, T, Stuwe, E, Hoelz, A. | | Deposit date: | 2012-12-05 | | Release date: | 2013-01-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and Functional Analysis of the C-Terminal Domain of Nup358/RanBP2.
J.Mol.Biol., 425, 2013
|
|
2GWC
 
 | | Crystal structure of plant glutamate cysteine ligase in complex with a transition state analogue | | Descriptor: | (2S)-2-amino-4-(S-butylsulfonimidoyl)butanoic acid, Glutamate cysteine ligase, MAGNESIUM ION | | Authors: | Hothorn, M, Wachter, A, Gromes, R, Stuwe, T, Rausch, T, Scheffzek, K. | | Deposit date: | 2006-05-04 | | Release date: | 2006-06-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structural basis for the redox control of plant glutamate cysteine ligase.
J.Biol.Chem., 281, 2006
|
|
2GWD
 
 | | Crystal structure of plant glutamate cysteine ligase in complex with Mg2+ and L-glutamate | | Descriptor: | ACETATE ION, GLUTAMIC ACID, Glutamate cysteine ligase, ... | | Authors: | Hothorn, M, Wachter, A, Gromes, R, Stuwe, T, Rausch, T, Scheffzek, K. | | Deposit date: | 2006-05-04 | | Release date: | 2006-06-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural basis for the redox control of plant glutamate cysteine ligase.
J.Biol.Chem., 281, 2006
|
|
4GA1
 
 | |
4GA0
 
 | |
4GA2
 
 | |
5HB7
 
 | |
5HB8
 
 | |
5HB5
 
 | |
