3MSL
 
 | |
9FEJ
 
 | | Structure of the RNA-dependent RNA polymerase P2 from the bacteriophage Phi8 | | Descriptor: | CALCIUM ION, RNA-directed RNA polymerase | | Authors: | Latimer-Smith, M, Salgado, P.S, Forsyth, I, Makeyev, E, Poranen, M, Stuart, D.I, Grimes, J.M, El Omari, K. | | Deposit date: | 2024-05-20 | | Release date: | 2024-10-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the RNA-dependent RNA polymerase P2 from the cystovirus phi 8.
Sci Rep, 14, 2024
|
|
9CYO
 
 | | Crystal structure of wild-type human PTP1B (PTPN1) at room temperature (298 K) | | Descriptor: | BETA-MERCAPTOETHANOL, MAGNESIUM ION, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Ebrahim, A, Perdikari, A, Woods, V.A, Lawler, K, Bounds, R, Singh, N.I, Mehlman, T, Riley, B.T, Sharma, S, Morris, J.W, Keogh, J.M, Henning, E, Smith, M, Farooqi, I.S, Keedy, D.A. | | Deposit date: | 2024-08-02 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structures of human PTP1B variants reveal allosteric sites to target for weight loss therapy.
Biorxiv, 2024
|
|
9CYQ
 
 | | Crystal structure of Q78R mutant human PTP1B (PTPN1) at room temperature (298 K) | | Descriptor: | BETA-MERCAPTOETHANOL, MAGNESIUM ION, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Ebrahim, A, Perdikari, A, Woods, V.A, Lawler, K, Bounds, R, Singh, N.I, Mehlman, T, Riley, B.T, Sharma, S, Morris, J.W, Keogh, J.M, Henning, E, Smith, M, Farooqi, I.S, Keedy, D.A. | | Deposit date: | 2024-08-02 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of human PTP1B variants reveal allosteric sites to target for weight loss therapy.
Biorxiv, 2024
|
|
9CYR
 
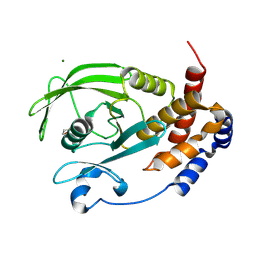 | | Crystal structure of D245G mutant human PTP1B (PTPN1) at room temperature (298 K) | | Descriptor: | BETA-MERCAPTOETHANOL, MAGNESIUM ION, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Ebrahim, A, Perdikari, A, Woods, V.A, Lawler, K, Bounds, R, Singh, N.I, Mehlman, T, Riley, B.T, Sharma, S, Morris, J.W, Keogh, J.M, Henning, E, Smith, M, Farooqi, I.S, Keedy, D.A. | | Deposit date: | 2024-08-02 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structures of human PTP1B variants reveal allosteric sites to target for weight loss therapy.
Biorxiv, 2024
|
|
9CYP
 
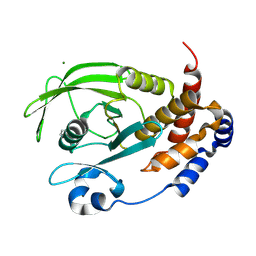 | | Crystal structure of I19V mutant human PTP1B (PTPN1) at room temperature (298 K) | | Descriptor: | BETA-MERCAPTOETHANOL, MAGNESIUM ION, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Ebrahim, A, Perdikari, A, Woods, V.A, Lawler, K, Bounds, R, Singh, N.I, Mehlman, T, Riley, B.T, Sharma, S, Morris, J.W, Keogh, J.M, Henning, E, Smith, M, Farooqi, I.S, Keedy, D.A. | | Deposit date: | 2024-08-02 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structures of human PTP1B variants reveal allosteric sites to target for weight loss therapy.
Biorxiv, 2024
|
|
4JXI
 
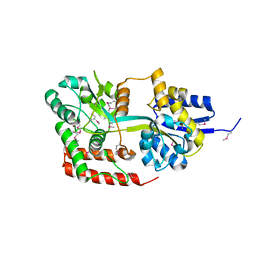 | | Directed evolution and rational design of a de novo designed esterase toward improved catalysis. Northeast Structural Genomics Consortium (NESG) Target OR184 | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, BROMIDE ION, GLYCEROL, ... | | Authors: | Kuzin, A, Smith, M.D, Richter, F, Lew, S, Seetharaman, R, Bryan, C, Lech, Z, Kiss, G, Moretti, R, Maglaqui, M, Xiao, R, Kohan, E, Smith, M, Everett, J.K, Nguyen, R, Pande, V, Hilvert, D, Kornhaber, G, Baker, D, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-03-28 | | Release date: | 2013-05-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Directed evolution and rational design of a de novo designed esterase toward improved catalysis. Northeast Structural Genomics Consortium (NESG) Target OR184
To be Published
|
|
6UNT
 
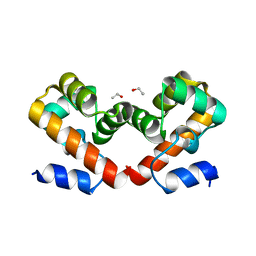 | | Barrier-to-autointegration factor soaked in DMSO: 1 of 14 in MSCS set | | Descriptor: | Barrier-to-autointegration factor, ETHANOL | | Authors: | Agarwal, S, Smith, M, De La Rosa, I, Kliment, A.V, Swartz, P, Segura-Totten, M, Mattos, C. | | Deposit date: | 2019-10-13 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Development of a structure-analysis pipeline using multiple-solvent crystal structures of barrier-to-autointegration factor.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6URE
 
 | | Barrier-to-autointegration factor Aqueous: 1 of 14 in MSCS set | | Descriptor: | Barrier-to-autointegration factor, ETHANOL | | Authors: | Agarwal, S, Smith, M, De La Rosa, I, Kliment, A.V, Swartz, P, Segura-Totten, M, Mattos, C. | | Deposit date: | 2019-10-23 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.653 Å) | | Cite: | Development of a structure-analysis pipeline using multiple-solvent crystal structures of barrier-to-autointegration factor.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6URK
 
 | | Barrier-to-autointegration factor soaked in Glycerol: 1 of 14 in MSCS set | | Descriptor: | Barrier-to-autointegration factor, ETHANOL | | Authors: | Agarwal, S, Smith, M, De La Rosa, I, Kliment, A.V, Swartz, P, Segura-Totten, M, Mattos, C. | | Deposit date: | 2019-10-23 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Development of a structure-analysis pipeline using multiple-solvent crystal structures of barrier-to-autointegration factor.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6USD
 
 | | Barrier-to-autointegration factor soaked in ethanol: 1 of 14 in MSCS set | | Descriptor: | Barrier-to-autointegration factor, ETHANOL | | Authors: | Agarwal, S, Smith, M, De La Rosa, I, Kliment, A.V, Swartz, P, Segura-Totten, M, Mattos, C. | | Deposit date: | 2019-10-25 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.653 Å) | | Cite: | Development of a structure-analysis pipeline using multiple-solvent crystal structures of barrier-to-autointegration factor.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6URJ
 
 | | Barrier-to-autointegration factor soaked in Acetone: 1 of 14 in MSCS set | | Descriptor: | Barrier-to-autointegration factor, ETHANOL | | Authors: | Agarwal, S, Smith, M, De La Rosa, I, Kliment, A.V, Swartz, P, Segura-Totten, M, Mattos, C. | | Deposit date: | 2019-10-23 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.653 Å) | | Cite: | Development of a structure-analysis pipeline using multiple-solvent crystal structures of barrier-to-autointegration factor.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6USB
 
 | | Barrier-to-autointegration factor soaked in urea: 1 of 14 in MSCS set | | Descriptor: | Barrier-to-autointegration factor, ETHANOL | | Authors: | Agarwal, S, Smith, M, De La Rosa, I, Kliment, A.V, Swartz, P, Segura-Totten, M, Mattos, C. | | Deposit date: | 2019-10-25 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Development of a structure-analysis pipeline using multiple-solvent crystal structures of barrier-to-autointegration factor.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6URL
 
 | | Barrier-to-autointegration factor soaked in isopropanol: 1 of 14 in MSCS set | | Descriptor: | Barrier-to-autointegration factor, ETHANOL | | Authors: | Agarwal, S, Smith, M, De La Rosa, I, Kliment, A.V, Swartz, P, Segura-Totten, M, Mattos, C. | | Deposit date: | 2019-10-23 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Development of a structure-analysis pipeline using multiple-solvent crystal structures of barrier-to-autointegration factor.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6URR
 
 | | Barrier-to-autointegration factor soaked in Dioxane: 1 of 14 in MSCS set | | Descriptor: | Barrier-to-autointegration factor, ETHANOL | | Authors: | Agarwal, S, Smith, M, De La Rosa, I, Kliment, A.V, Swartz, P, Segura-Totten, M, Mattos, C. | | Deposit date: | 2019-10-24 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Development of a structure-analysis pipeline using multiple-solvent crystal structures of barrier-to-autointegration factor.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6US1
 
 | | Barrier-to-autointegration factor soaked in isobutanol: 1 of 14 in MSCS set | | Descriptor: | Barrier-to-autointegration factor, ETHANOL | | Authors: | Agarwal, S, Smith, M, De La Rosa, I, Kliment, A.V, Swartz, P, Segura-Totten, M, Mattos, C. | | Deposit date: | 2019-10-24 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.653 Å) | | Cite: | Development of a structure-analysis pipeline using multiple-solvent crystal structures of barrier-to-autointegration factor.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6US7
 
 | | Barrier-to-autointegration factor soaked in TMAO: 1 of 14 in MSCS set | | Descriptor: | Barrier-to-autointegration factor, ETHANOL | | Authors: | Agarwal, S, Smith, M, De La Rosa, I, Kliment, A.V, Swartz, P, Segura-Totten, M, Mattos, C. | | Deposit date: | 2019-10-24 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.653 Å) | | Cite: | Development of a structure-analysis pipeline using multiple-solvent crystal structures of barrier-to-autointegration factor.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6USI
 
 | | Barrier-to-autointegration factor soaked in 1,6-hexanediol: 1 of 14 in MSCS set | | Descriptor: | Barrier-to-autointegration factor, ETHANOL | | Authors: | Agarwal, S, Smith, M, De La Rosa, I, Kliment, A.V, Swartz, P, Segura-Totten, M, Mattos, C. | | Deposit date: | 2019-10-26 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.653 Å) | | Cite: | Development of a structure-analysis pipeline using multiple-solvent crystal structures of barrier-to-autointegration factor.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6URZ
 
 | | Barrier-to-autointegration factor soaked in methanol: 1 of 14 in MSCS set | | Descriptor: | Barrier-to-autointegration factor, ETHANOL, METHANOL | | Authors: | Agarwal, S, Smith, M, De La Rosa, I, Kliment, A.V, Swartz, P, Segura-Totten, M, Mattos, C. | | Deposit date: | 2019-10-24 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.653 Å) | | Cite: | Development of a structure-analysis pipeline using multiple-solvent crystal structures of barrier-to-autointegration factor.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6URN
 
 | | Barrier-to-autointegration factor t-butanol: 1 of 14 in MSCS set | | Descriptor: | Barrier-to-autointegration factor, ETHANOL | | Authors: | Agarwal, S, Smith, M, De La Rosa, I, Kliment, A.V, Swartz, P, Segura-Totten, M, Mattos, C. | | Deposit date: | 2019-10-23 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Development of a structure-analysis pipeline using multiple-solvent crystal structures of barrier-to-autointegration factor.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6US0
 
 | | Barrier-to-autointegration factor soaked in R,S,R-bisfuranol (RSR): 1 of 14 in MSCS set | | Descriptor: | Barrier-to-autointegration factor, ETHANOL | | Authors: | Agarwal, S, Smith, M, De La Rosa, I, Kliment, A.V, Swartz, P, Segura-Totten, M, Mattos, C. | | Deposit date: | 2019-10-24 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.653 Å) | | Cite: | Development of a structure-analysis pipeline using multiple-solvent crystal structures of barrier-to-autointegration factor.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
1PIH
 
 | | THE THREE DIMENSIONAL STRUCTURE OF THE PARAMAGNETIC PROTEIN HIPIP I FROM E.HALOPHILA THROUGH NUCLEAR MAGNETIC RESONANCE | | Descriptor: | HIGH POTENTIAL IRON SULFUR PROTEIN, IRON/SULFUR CLUSTER | | Authors: | Banci, L, Bertini, I, Eltis, L.D, Felli, I, Kastrau, D.H.W, Luchinat, C, Piccioli, M, Pierattelli, R, Smith, M. | | Deposit date: | 1994-08-03 | | Release date: | 1994-12-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional structure in solution of the paramagnetic high-potential iron-sulfur protein I from Ectothiorhodospira halophila through nuclear magnetic resonance.
Eur.J.Biochem., 225, 1994
|
|
1PIJ
 
 | | THE THREE DIMENSIONAL STRUCTURE OF THE PARAMAGNETIC PROTEIN HIPIP I FROM E.HALOPHILA THROUGH NUCLEAR MAGNETIC RESONANCE | | Descriptor: | HIGH POTENTIAL IRON SULFUR PROTEIN, IRON/SULFUR CLUSTER | | Authors: | Banci, L, Bertini, I, Eltis, L.D, Felli, I.C, Kastrau, D.H.W, Luchinat, C, Piccioli, M, Pierattelli, R, Smith, M. | | Deposit date: | 1994-11-11 | | Release date: | 1995-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional structure in solution of the paramagnetic high-potential iron-sulfur protein I from Ectothiorhodospira halophila through nuclear magnetic resonance.
Eur.J.Biochem., 225, 1994
|
|
1NZ2
 
 | | K45E Variant of Horse Heart Myoglobin | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Hunter, C.L, Maurus, R, Mauk, M.R, Lee, H, Raven, E.L, Tong, H, Nguyen, N, Smith, M, Brayer, G.D, Mauk, A.G. | | Deposit date: | 2003-02-15 | | Release date: | 2003-04-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Introduction and characterization of a functionally linked metal ion binding site at
the exposed heme edge of myoglobin
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
3EV3
 
 | | Crystal Structure of Ribonuclease A in 70% t-Butanol | | Descriptor: | Ribonuclease pancreatic, TERTIARY-BUTYL ALCOHOL | | Authors: | Dechene, M, Wink, G, Smith, M, Swartz, P, Mattos, C. | | Deposit date: | 2008-10-12 | | Release date: | 2009-06-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Multiple solvent crystal structures of ribonuclease A: An assessment of the method
Proteins, 76, 2009
|
|
