2W8I
 
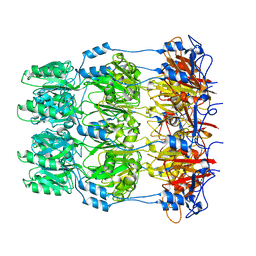 | | Crystal structure of Wza24-345. | | Descriptor: | PUTATIVE OUTER MEMBRANE LIPOPROTEIN WZA | | Authors: | Hagelueken, G, Ingledew, W.J, Huang, H, Petrovic-Stojanovska, B, Whitfield, C, ElMkami, H, Schiemann, O, Naismith, J.H. | | Deposit date: | 2009-01-16 | | Release date: | 2009-02-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Peldor Distance Fingerprinting of the Octameric Outer-Membrane Protein Wza from Escherichia Coli.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
2W8H
 
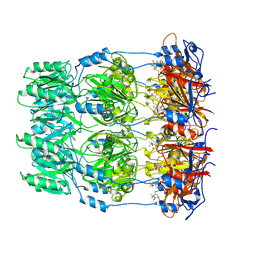 | | Crystal structure of spin labeled Wza24-345. | | Descriptor: | CHLORIDE ION, PUTATIVE OUTER MEMBRANE LIPOPROTEIN WZA, S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate | | Authors: | Hagelueken, G, Ingledew, W.J, Huang, H, Petrovic-Stojanovska, B, Whitfield, C, ElMkami, H, Schiemann, O, Naismith, J.H. | | Deposit date: | 2009-01-16 | | Release date: | 2009-02-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Peldor Distance Fingerprinting of the Octameric Outer-Membrane Protein Wza from Escherichia Coli.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
7PDU
 
 | |
5FH9
 
 | |
3OT0
 
 | |
7APM
 
 | | tRNA-guanine transglycosylase H319C mutant spin-labeled with MTSL. | | Descriptor: | CHLORIDE ION, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Nguyen, D, Heine, A, Klebe, G. | | Deposit date: | 2020-10-18 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Unraveling a Ligand-Induced Twist of a Homodimeric Enzyme by Pulsed Electron-Electron Double Resonance.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7APL
 
 | |
5OFR
 
 | |
5OFP
 
 | |
4AGF
 
 | |
4AGE
 
 | |
4BWW
 
 | | Crystal structure of spin labelled azurin T21R1. | | Descriptor: | AZURIN, COPPER (II) ION, GLYCEROL, ... | | Authors: | Hagelueken, G. | | Deposit date: | 2013-07-04 | | Release date: | 2014-06-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | High-Resolution Crystal Structure of Spin Labelled (T21R1) Azurin from Pseudomonas Aeruginosa: A Challenging Structural Benchmark for in Silico Spin Labelling Algorithms.
Bmc Struct.Biol., 14, 2014
|
|
5I26
 
 | | Azurin T30R1, crystal form I | | Descriptor: | Azurin, COPPER (II) ION | | Authors: | Hagelueken, G. | | Deposit date: | 2016-02-08 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.888 Å) | | Cite: | Determination of nitroxide spin label conformations via PELDOR and X-ray crystallography.
Phys Chem Chem Phys, 18, 2016
|
|
5I28
 
 | | Azurin T30R1, crystal form II | | Descriptor: | Azurin, COPPER (II) ION, GLYCEROL | | Authors: | Hagelueken, G. | | Deposit date: | 2016-02-08 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Determination of nitroxide spin label conformations via PELDOR and X-ray crystallography.
Phys Chem Chem Phys, 18, 2016
|
|
