3GNA
 
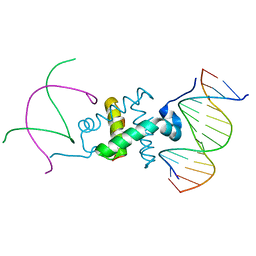 | | Crystal structure of the RAG1 nonamer-binding domain with DNA | | Descriptor: | 5'-D(*AP*CP*TP*TP*AP*AP*CP*AP*AP*AP*AP*AP*CP*C)-3', 5'-D(*TP*GP*GP*TP*TP*TP*TP*TP*GP*TP*TP*AP*AP*G)-3', V(D)J recombination-activating protein 1 | | Authors: | Yin, F.F, Bailey, S, Innis, C.A, Steitz, T.A, Schatz, D.G. | | Deposit date: | 2009-03-16 | | Release date: | 2009-04-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the RAG1 nonamer binding domain with DNA reveals a dimer that mediates DNA synapsis.
Nat.Struct.Mol.Biol., 16, 2009
|
|
3GNB
 
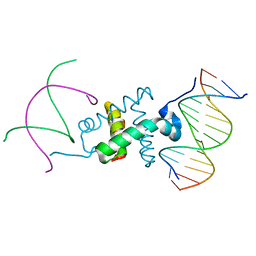 | | Crystal structure of the RAG1 nonamer-binding domain with DNA | | Descriptor: | 5'-D(*AP*AP*TP*TP*TP*TP*CP*AP*GP*AP*AP*AP*CP*C)-3', 5'-D(*AP*GP*GP*TP*TP*TP*CP*TP*GP*AP*AP*AP*AP*C)-3', V(D)J recombination-activating protein 1 | | Authors: | Yin, F.F, Bailey, S, Innis, C.A, Steitz, T.A, Schatz, D.G. | | Deposit date: | 2009-03-16 | | Release date: | 2009-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the RAG1 nonamer binding domain with DNA reveals a dimer that mediates DNA synapsis.
Nat.Struct.Mol.Biol., 16, 2009
|
|
1RMD
 
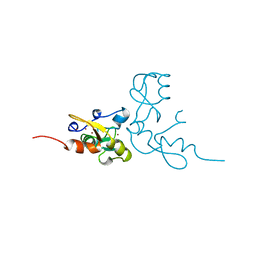 | | RAG1 DIMERIZATION DOMAIN | | Descriptor: | RAG1, ZINC ION | | Authors: | Bellon, S.F, Rodgers, K.K, Schatz, D.G, Coleman, J.E, Steitz, T.A. | | Deposit date: | 1997-01-10 | | Release date: | 1997-07-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the RAG1 dimerization domain reveals multiple zinc-binding motifs including a novel zinc binuclear cluster.
Nat.Struct.Biol., 4, 1997
|
|
6PQY
 
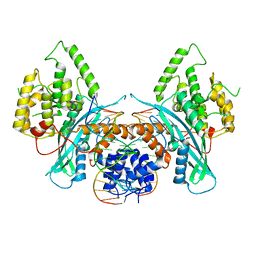 | | Cryo-EM structure of HzTransib/TIR DNA transposon end complex (TEC) | | Descriptor: | DNA (5'-D(P*CP*AP*CP*GP*GP*TP*GP*GP*AP*TP*CP*GP*AP*AP*AP*A)-3'), DNA (5'-D(P*TP*TP*TP*TP*CP*GP*AP*TP*CP*CP*AP*CP*CP*GP*TP*G)-3'), Putative DNA-mediated transposase | | Authors: | Liu, C, Yang, Y, Schatz, D.G. | | Deposit date: | 2019-07-10 | | Release date: | 2019-10-09 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structures of a RAG-like transposase during cut-and-paste transposition.
Nature, 575, 2019
|
|
6PR5
 
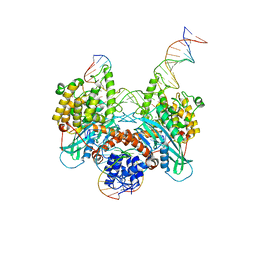 | | Cryo-EM structure of HzTransib strand transfer complex (STC) | | Descriptor: | DNA (30-MER), DNA (39-MER), DNA (5'-D(*GP*AP*TP*CP*TP*GP*GP*CP*CP*TP*AP*GP*AP*TP*CP*TP*CP*A)-3'), ... | | Authors: | Liu, C, Yang, Y, Schatz, D.G. | | Deposit date: | 2019-07-10 | | Release date: | 2019-10-09 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures of a RAG-like transposase during cut-and-paste transposition.
Nature, 575, 2019
|
|
6PQN
 
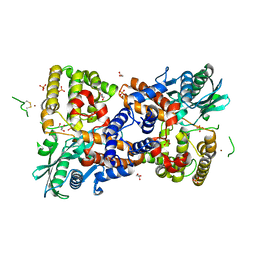 | | Crystal structure of HzTransib transposase | | Descriptor: | GLYCEROL, PHOSPHATE ION, Putative DNA-mediated transposase, ... | | Authors: | Liu, C, Yang, Y, Schatz, D.G. | | Deposit date: | 2019-07-09 | | Release date: | 2019-10-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structures of a RAG-like transposase during cut-and-paste transposition.
Nature, 575, 2019
|
|
6PQX
 
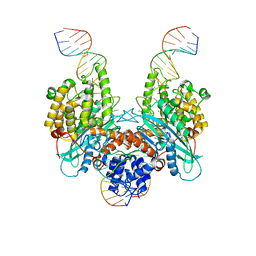 | | Cryo-EM structure of HzTransib/nicked TIR substrate DNA hairpin forming complex (HFC) | | Descriptor: | CALCIUM ION, DNA (5'-D(P*CP*AP*CP*GP*GP*TP*GP*GP*AP*TP*CP*GP*AP*AP*AP*A)-3'), DNA (5'-D(P*TP*CP*TP*GP*GP*CP*CP*TP*AP*GP*AP*TP*CP*T)-3'), ... | | Authors: | Liu, C, Yang, Y, Schatz, D.G. | | Deposit date: | 2019-07-10 | | Release date: | 2019-10-09 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structures of a RAG-like transposase during cut-and-paste transposition.
Nature, 575, 2019
|
|
6PQU
 
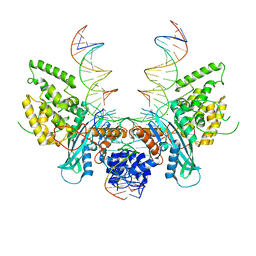 | | Cryo-EM structure of HzTransib/nicked TIR substrate DNA pre-reaction complex (PRC) | | Descriptor: | DNA (5'-D(P*AP*TP*CP*TP*GP*GP*CP*CP*TP*AP*GP*AP*TP*CP*T)-3'), DNA (5'-D(P*CP*AP*CP*GP*GP*TP*GP*GP*AP*TP*CP*GP*AP*AP*AP*A)-3'), DNA-mediated transposase, ... | | Authors: | Liu, C, Yang, Y, Schatz, D.G. | | Deposit date: | 2019-07-10 | | Release date: | 2019-10-09 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures of a RAG-like transposase during cut-and-paste transposition.
Nature, 575, 2019
|
|
6PQR
 
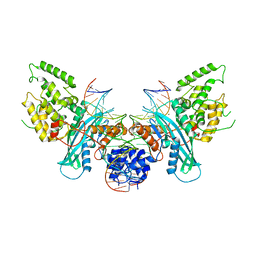 | | Cryo-EM structure of HzTransib/intact TIR substrate DNA pre-reaction complex (PRC) | | Descriptor: | DNA (5'-D(*CP*TP*AP*GP*AP*TP*CP*TP*CP*AP*CP*GP*GP*TP*GP*GP*AP*TP*CP*GP*AP*AP*AP*A)-3'), DNA (5'-D(P*TP*TP*TP*TP*CP*GP*AP*TP*CP*CP*AP*CP*CP*GP*TP*GP*AP*GP*AP*TP*CP*TP*AP*G)-3'), DNA-mediated transposase, ... | | Authors: | Liu, C, Yang, Y, Schatz, D.G. | | Deposit date: | 2019-07-09 | | Release date: | 2019-10-09 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of a RAG-like transposase during cut-and-paste transposition.
Nature, 575, 2019
|
|
6XNX
 
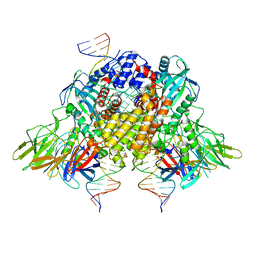 | | Structure of RAG1 (R848M/E649V)-RAG2-DNA Strand Transfer Complex (Dynamic-Form) | | Descriptor: | 12RSS integration strand DNA (55-MER), 12RSS signal top strand DNA (34-MER), 23RSS integration strand DNA (66-MER), ... | | Authors: | Zhang, Y, Corbett, E, Wu, S, Schatz, D.G. | | Deposit date: | 2020-07-05 | | Release date: | 2020-08-26 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis for the activation and suppression of transposition during evolution of the RAG recombinase.
Embo J., 39, 2020
|
|
6XNZ
 
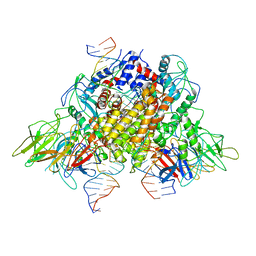 | | Structure of RAG1 (R848M/E649V)-RAG2-DNA Target Capture Complex | | Descriptor: | 12RSS integration strand (34-mer), 12RSS non-integration strand (34-mer), 23RSS integration strand (45-mer), ... | | Authors: | Zhang, Y, Corbett, E, Wu, S, Schatz, D.G. | | Deposit date: | 2020-07-05 | | Release date: | 2020-08-26 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for the activation and suppression of transposition during evolution of the RAG recombinase.
Embo J., 39, 2020
|
|
6XNY
 
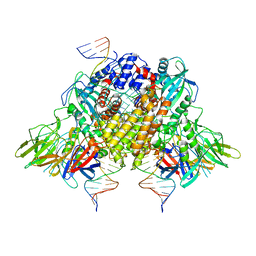 | | Structure of RAG1 (R848M/E649V)-RAG2-DNA Strand Transfer Complex (Paired-Form) | | Descriptor: | 12RSS integration strand (55-mer), 12RSS signal DNA top strand (34-mer), 23RSS integration strand (66-mer), ... | | Authors: | Zhang, Y, Corbett, E, Wu, S, Schatz, D.G. | | Deposit date: | 2020-07-05 | | Release date: | 2020-08-26 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for the activation and suppression of transposition during evolution of the RAG recombinase.
Embo J., 39, 2020
|
|
6B40
 
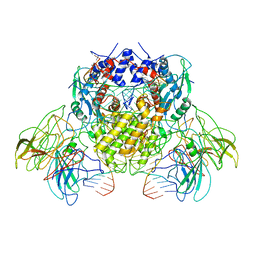 | | BbRAGL-3'TIR synaptic complex with nicked DNA refined with C2 symmetry | | Descriptor: | 31TIR intact strand, 31TIR pre-nicked strand of flanking DNA, 31TIR pre-nicked strand of signal DNA, ... | | Authors: | Zhang, Y, Cheng, T.C, Xiong, Y, Schatz, D.G. | | Deposit date: | 2017-09-25 | | Release date: | 2019-03-20 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Transposon molecular domestication and the evolution of the RAG recombinase.
Nature, 569, 2019
|
|
7N0D
 
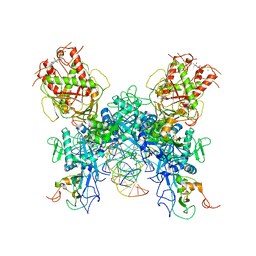 | |
7N0B
 
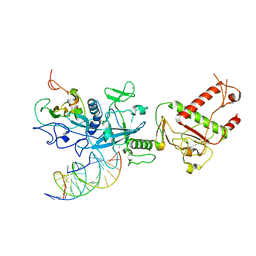 | | Cryo-EM structure of SARS-CoV-2 nsp10-nsp14 (WT)-RNA complex | | Descriptor: | CALCIUM ION, Non-structural protein 10, Proofreading exoribonuclease, ... | | Authors: | Liu, C, Yang, Y. | | Deposit date: | 2021-05-25 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis of mismatch recognition by a SARS-CoV-2 proofreading enzyme.
Science, 373, 2021
|
|
7N0C
 
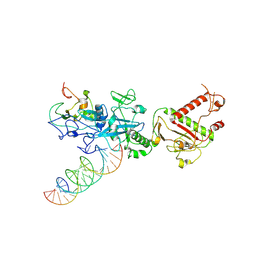 | |
6WL5
 
 | | Crystal structure of EcmrR C-terminal domain | | Descriptor: | 1,2-ETHANEDIOL, CETYL-TRIMETHYL-AMMONIUM, CHLORIDE ION, ... | | Authors: | Yang, Y, Liu, C, Liu, B. | | Deposit date: | 2020-04-18 | | Release date: | 2021-04-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural visualization of transcription activated by a multidrug-sensing MerR family regulator.
Nat Commun, 12, 2021
|
|
6XLK
 
 | | Cryo-EM structure of EcmrR-DNA complex in EcmrR-RPitc-4nt | | Descriptor: | CHAPSO, MerR family transcriptional regulator EcmrR, TETRAPHENYLANTIMONIUM ION, ... | | Authors: | Yang, Y, Liu, C, Liu, B. | | Deposit date: | 2020-06-28 | | Release date: | 2021-04-07 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural visualization of transcription activated by a multidrug-sensing MerR family regulator.
Nat Commun, 12, 2021
|
|
6XLA
 
 | | Cryo-EM structure of EcmrR-DNA complex in EcmrR-RPitc-3nt | | Descriptor: | MerR family transcriptional regulator EcmrR, TETRAPHENYLANTIMONIUM ION, synthetic non-template strand DNA (54-MER), ... | | Authors: | Yang, Y, Liu, C, Shi, W, Liu, B. | | Deposit date: | 2020-06-28 | | Release date: | 2021-04-07 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural visualization of transcription activated by a multidrug-sensing MerR family regulator.
Nat Commun, 12, 2021
|
|
6XL6
 
 | | Cryo-EM structure of EcmrR-DNA complex in EcmrR-RPo | | Descriptor: | CHAPSO, MerR family transcriptional regulator EcmrR, TETRAPHENYLANTIMONIUM ION, ... | | Authors: | Yang, Y, Liu, C, Liu, B. | | Deposit date: | 2020-06-28 | | Release date: | 2021-04-07 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural visualization of transcription activated by a multidrug-sensing MerR family regulator.
Nat Commun, 12, 2021
|
|
6XLN
 
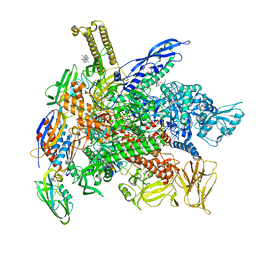 | | Cryo-EM structure of E. coli RNAP-DNA elongation complex 2 (RDe2) in EcmrR-dependent transcription | | Descriptor: | 9-nt RNA transcript, CHAPSO, DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Yang, Y, Liu, C, Liu, B. | | Deposit date: | 2020-06-28 | | Release date: | 2021-04-07 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural visualization of transcription activated by a multidrug-sensing MerR family regulator.
Nat Commun, 12, 2021
|
|
6XLM
 
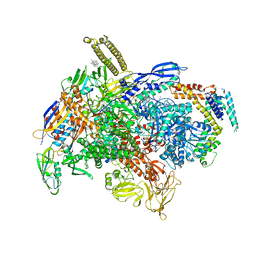 | | Cryo-EM structure of E.coli RNAP-DNA elongation complex 1 (RDe1) in EcmrR-dependent transcription | | Descriptor: | 9-nt RNA transcript, CHAPSO, DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Yang, Y, Liu, C, Liu, B. | | Deposit date: | 2020-06-28 | | Release date: | 2021-04-07 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural visualization of transcription activated by a multidrug-sensing MerR family regulator.
Nat Commun, 12, 2021
|
|
6XLL
 
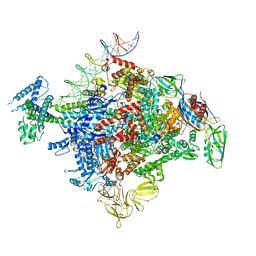 | | Cryo-EM structure of E. coli RNAP-promoter initial transcribing complex with 5-nt RNA transcript (RPitc-5nt) | | Descriptor: | CHAPSO, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Yang, Y, Liu, C, Liu, B. | | Deposit date: | 2020-06-28 | | Release date: | 2021-04-07 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural visualization of transcription activated by a multidrug-sensing MerR family regulator.
Nat Commun, 12, 2021
|
|
6XL9
 
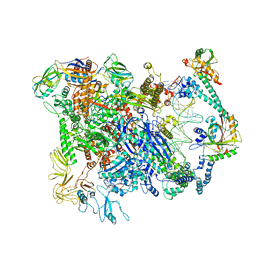 | | Cryo-EM structure of EcmrR-RNAP-promoter initial transcribing complex with 3-nt RNA transcript (EcmrR-RPitc-3nt) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Yang, Y, Liu, C, Shi, W, Liu, B. | | Deposit date: | 2020-06-28 | | Release date: | 2021-04-07 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural visualization of transcription activated by a multidrug-sensing MerR family regulator.
Nat Commun, 12, 2021
|
|
6XLJ
 
 | | Cryo-EM structure of EcmrR-RNAP-promoter initial transcribing complex with 4-nt RNA transcript (EcmrR-RPitc-4nt) | | Descriptor: | CHAPSO, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Yang, Y, Liu, C, Liu, B. | | Deposit date: | 2020-06-28 | | Release date: | 2021-04-07 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural visualization of transcription activated by a multidrug-sensing MerR family regulator.
Nat Commun, 12, 2021
|
|
