2GDQ
 
 | | Crystal structure of mandelate racemase/muconate lactonizing enzyme from Bacillus subtilis at 1.8 A resolution | | Descriptor: | yitF | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Schwinn, K.D, Emtage, S, Thompson, D.A, Rutter, M.E, Dickey, M, Groshong, C, Bain, K.T, Adams, J.M, Reyes, C, Rooney, I, Powell, A, Boice, A, Gheyi, T, Ozyurt, S, Atwell, S, Wasserman, S.R, Burley, S.K, Sali, A, Babbitt, P, Pieper, U, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-03-16 | | Release date: | 2006-04-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of mandelate racemase/muconate lactonizing enzyme from Bacillus subtilis at 1.8 A resolution
To be Published
|
|
2GGE
 
 | | Crystal Structure of Mandelate Racemase/Muconate Lactonizing Enzyme from Bacillus Subtilis complexed with MG++ at 1.8 A | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, yitF | | Authors: | Malashkevich, V.N, Sauder, J.M, Schwinn, K.D, Emtage, S, Thompson, D.A, Rutter, M.E, Dickey, M, Groshong, C, Bain, K.T, Adams, J.M, Reyes, C, Rooney, I, Powell, A, Boice, A, Gheyi, T, Ozyurt, S, Atwell, S, Wasserman, S.R, Burley, S.K, Sali, A, Babbitt, P, Pieper, U, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-03-23 | | Release date: | 2006-04-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal Structure of Mandelate Racemase/Muconate Lactonizing Enzyme from Bacillus Subtilis complexed with MG++ at 1.8 A
To be Published
|
|
2GL5
 
 | | Crystal Structure of Putative Dehydratase from Salmonella Thyphimurium | | Descriptor: | GLYCEROL, MAGNESIUM ION, putative dehydratase protein | | Authors: | Patskovsky, Y, Sauder, J.M, Dickey, M, Adams, J.M, Ozyurt, S, Wasserman, S.R, Gerlt, J, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-04-04 | | Release date: | 2006-04-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of Putative Dehydratase from Salmonella Thyphimurium Lt2
To be Published
|
|
2GSH
 
 | | Crystal structure of L-rhamnonate dehydratase from Salmonella typhimurium | | Descriptor: | GLYCEROL, L-RHAMNONATE DEHYDRATASE, MAGNESIUM ION | | Authors: | Patskovsky, Y, Malashkevich, V.N, Sauder, J.M, Dickey, M, Adams, J.M, Wasserman, S.R, Gerlt, J, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-04-26 | | Release date: | 2006-05-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.393 Å) | | Cite: | Crystal Structure of L-rhamnonate dehydratase from Salmonella Typhimurium Lt2
To be Published
|
|
8V9B
 
 | | Lipoprotein(a) Kringle IV domain 7 - Lp(a) KIV7 in complex with LY3441732 | | Descriptor: | (2S,2'S)-3,3'-[carbonylbis(azanediyl-3,1-phenylene)]bis{2-[(3R)-pyrrolidin-1-ium-3-yl]propanoate}, Apolipoprotein(a), MAGNESIUM ION, ... | | Authors: | Hendle, J, Weichert, K, Sauder, J.M. | | Deposit date: | 2023-12-07 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Discovery of potent small-molecule inhibitors of lipoprotein(a) formation.
Nature, 629, 2024
|
|
8V8Z
 
 | |
2RDM
 
 | | Crystal structure of response regulator receiver protein from Sinorhizobium medicae WSM419 | | Descriptor: | GLYCEROL, Response regulator receiver protein | | Authors: | Patskovsky, Y, Yan, Q, Zhan, C, Toro, R, Meyer, A.J, Gilmore, M, Hu, S, Groshong, C, Rodgers, L, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-09-24 | | Release date: | 2007-10-09 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of response regulator receiver protein from Sinorhizobium medicae WSM419.
To be Published
|
|
2RAG
 
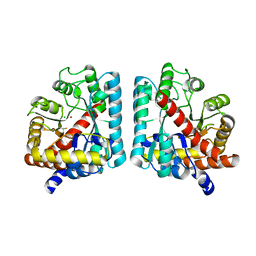 | | Crystal structure of aminohydrolase from Caulobacter crescentus | | Descriptor: | CHLORIDE ION, Dipeptidase, ZINC ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-09-14 | | Release date: | 2007-10-02 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of aminohydrolase from Caulobacter crescentus.
To be Published
|
|
2R9G
 
 | | Crystal structure of the C-terminal fragment of AAA ATPase from Enterococcus faecium | | Descriptor: | AAA ATPase, central region, ACETATE ION, ... | | Authors: | Ramagopal, U.A, Patskovsky, Y, Bonanno, J.B, Shi, W, Toro, R, Meyer, A.J, Rutter, M, Wu, B, Groshong, C, Gheyi, T, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-09-12 | | Release date: | 2007-10-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal Structure of the C-Terminal Domain of AAA ATPase from Enterococcus faecium.
To be Published
|
|
2R5X
 
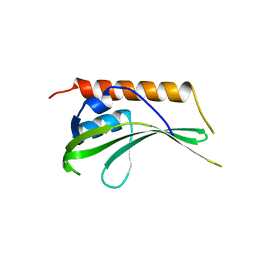 | | Crystal structure of uncharacterized conserved protein YugN from Geobacillus kaustophilus HTA426 | | Descriptor: | Uncharacterized conserved protein | | Authors: | Ramagopal, U.A, Patskovsky, Y, Shi, W, Toro, R, Meyer, A.J, Freeman, J, Wu, B, Koss, J, Groshong, C, Rodgers, L, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-09-04 | | Release date: | 2007-09-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystal structure of uncharacterized protein YugN from Geobacillus kaustophilus HTA426.
To be Published
|
|
2RG4
 
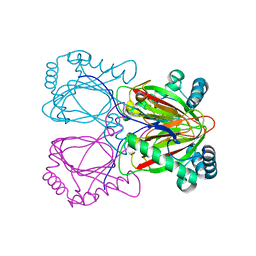 | | Crystal structure of the uncharacterized protein Q2CBJ1_9RHOB from Oceanicola granulosus HTCC2516 | | Descriptor: | FE (III) ION, Uncharacterized protein | | Authors: | Malashkevich, V.N, Toro, R, Meyer, A.J, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-10-02 | | Release date: | 2007-10-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the uncharacterized protein Q2CBJ1_9RHOB from Oceanicola granulosus HTCC2516.
To be Published
|
|
2RBB
 
 | |
2RDX
 
 | | Crystal structure of mandelate racemase/muconate lactonizing enzyme from Roseovarius nubinhibens ISM | | Descriptor: | GLYCEROL, MAGNESIUM ION, Mandelate racemase/muconate lactonizing enzyme, ... | | Authors: | Patskovsky, Y, Bonanno, J, Sauder, J.M, Ozyurt, S, Gilmore, M, Lau, C, Maletic, M, Gheyi, T, Wasserman, S.R, Koss, J, Gerlt, J.A, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-09-25 | | Release date: | 2007-10-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of mandelate racemase/muconate lactonizing enzyme from Roseovarius nubinhibens ISM.
To be Published
|
|
2RJN
 
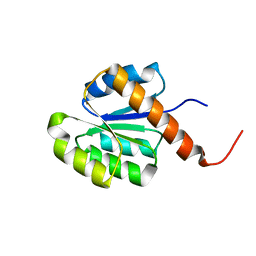 | | Crystal structure of an uncharacterized protein Q2BKU2 from Neptuniibacter caesariensis | | Descriptor: | Response regulator receiver:Metal-dependent phosphohydrolase, HD subdomain | | Authors: | Malashkevich, V.N, Toro, R, Meyer, A.J, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-10-15 | | Release date: | 2007-10-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of an uncharacterized protein Q2BKU2 from Neptuniibacter caesariensis.
To be Published
|
|
3LTE
 
 | | CRYSTAL STRUCTURE OF RESPONSE REGULATOR (SIGNAL RECEIVER DOMAIN) FROM Bermanella marisrubri | | Descriptor: | GLYCEROL, PHOSPHATE ION, Response regulator | | Authors: | Patskovsky, Y, Toro, R, Gilmore, M, Miller, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-02-15 | | Release date: | 2010-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CRYSTAL STRUCTURE OF RESPONSE REGULATOR SIGNAL RECEIVER DOMAIN FROM Bermanella marisrubri RED65
To be Published
|
|
3ME7
 
 | | Crystal structure of putative electron transport protein aq_2194 from Aquifex aeolicus VF5 | | Descriptor: | Putative uncharacterized protein, SULFATE ION | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-03-31 | | Release date: | 2010-04-14 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of putative electron transport protein aq_2194 from Aquifex aeolicus VF5
To be Published
|
|
3MKV
 
 | | Crystal structure of amidohydrolase eaj56179 | | Descriptor: | CARBONATE ION, GLYCEROL, PUTATIVE AMIDOHYDROLASE, ... | | Authors: | Patskovsky, Y, Bonanno, J, Ozyurt, S, Sauder, J.M, Freeman, J, Wu, B, Smith, D, Bain, K, Rodgers, L, Wasserman, S.R, Raushel, F.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-04-15 | | Release date: | 2010-04-28 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Functional identification and structure determination of two novel prolidases from cog1228 in the amidohydrolase superfamily .
Biochemistry, 49, 2010
|
|
3MAE
 
 | | CRYSTAL STRUCTURE OF PROBABLE DIHYDROLIPOAMIDE ACETYLTRANSFERASE FROM LISTERIA MONOCYTOGENES 4b F2365 | | Descriptor: | 2-oxoisovalerate dehydrogenase E2 component, dihydrolipoamide acetyltransferase, CHLORIDE ION, ... | | Authors: | Patskovsky, Y, Toro, R, Gilmore, M, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-03-23 | | Release date: | 2010-04-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | CRYSTAL STRUCTURE OF A CATALYTIC DOMAIN OF DIHYDROLIPOAMIDE ACETYLTRANSFERASE FROM LISTERIA MONOCYTOGENES 4b F2365
To be Published
|
|
3ME8
 
 | | Crystal structure of putative electron transfer protein aq_2194 from Aquifex aeolicus VF5 | | Descriptor: | Putative uncharacterized protein | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-03-31 | | Release date: | 2010-04-14 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of putative electron transfer protein aq_2194 from Aquifex aeolicus VF5
To be Published
|
|
3MGK
 
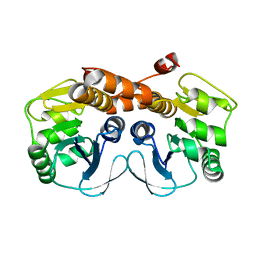 | | CRYSTAL STRUCTURE OF PROBABLE PROTEASE/AMIDASE FROM Clostridium acetobutylicum ATCC 824 | | Descriptor: | Intracellular protease/amidase related enzyme (ThiJ family) | | Authors: | Patskovsky, Y, Toro, R, Freeman, J, Iizuka, M, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-04-06 | | Release date: | 2010-04-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CRYSTAL STRUCTURE OF PROBABLE PROTEASE/AMIDASE FROM Clostridium acetobutylicum
To be Published
|
|
3MMZ
 
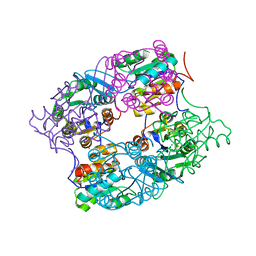 | | CRYSTAL STRUCTURE OF putative HAD family hydrolase from Streptomyces avermitilis MA-4680 | | Descriptor: | CALCIUM ION, CHLORIDE ION, putative HAD family hydrolase | | Authors: | Malashkevich, V.N, Ramagopal, U.A, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-04-20 | | Release date: | 2010-04-28 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural basis for the divergence of substrate specificity and biological function within HAD phosphatases in lipopolysaccharide and sialic acid biosynthesis.
Biochemistry, 52, 2013
|
|
3ME5
 
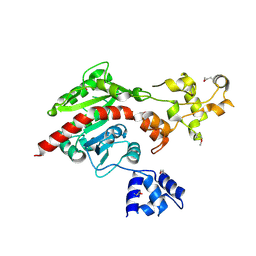 | | Crystal structure of putative dna cytosine methylase from shigella flexneri 2a str. 2457T | | Descriptor: | Cytosine-specific methyltransferase | | Authors: | Ramagopal, U.A, Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-03-31 | | Release date: | 2010-04-21 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of putative dna cytosine methylase from shigella flexneri 2a str. 2457T
To be Published
|
|
3CZ5
 
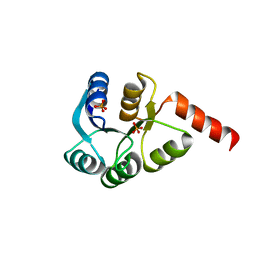 | | Crystal structure of two-component response regulator, LuxR family, from Aurantimonas sp. SI85-9A1 | | Descriptor: | PHOSPHATE ION, Two-component response regulator, LuxR family | | Authors: | Malashkevich, V.N, Toro, R, Wasserman, S.R, Meyer, A, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-04-28 | | Release date: | 2008-05-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of two-component response regulator, LuxR family, from Aurantimonas sp. SI85-9A1.
To be Published
|
|
3CJP
 
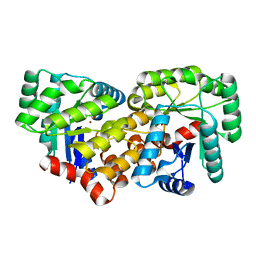 | | Crystal structure of an uncharacterized amidohydrolase CAC3332 from Clostridium acetobutylicum | | Descriptor: | Predicted amidohydrolase, dihydroorotase family, ZINC ION | | Authors: | Malashkevich, V.N, Toro, R, Ramagopal, U.A, Bonanno, J.B, Meyer, A, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-03-13 | | Release date: | 2008-03-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of an uncharacterized amidohydrolase CAC3332 from Clostridium acetobutylicum.
To be Published
|
|
3CG0
 
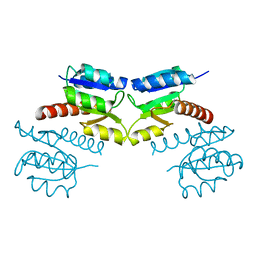 | | Crystal structure of signal receiver domain of modulated diguanylate cyclase from Desulfovibrio desulfuricans G20, an example of alternate folding | | Descriptor: | Response regulator receiver modulated diguanylate cyclase with PAS/PAC sensor | | Authors: | Patskovsky, Y, Bonanno, J.B, Romero, R, Gilmore, M, Chang, S, Groshong, C, Koss, J, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-03-04 | | Release date: | 2008-03-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of Signal Receiver Domain of Modulated Diguanylate Cyclase from Desulfovibrio desulfuricans.
To be Published
|
|
