5XJE
 
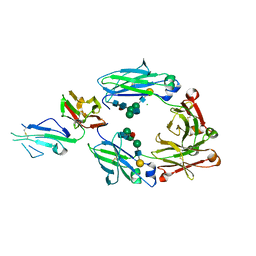 | | Crystal structure of fucosylated IgG1 Fc complexed with bis-glycosylated soluble form of Fc gamma receptor IIIa | | Descriptor: | CHLORIDE ION, Immunoglobulin gamma-1 heavy chain, Low affinity immunoglobulin gamma Fc region receptor III-A, ... | | Authors: | Sakae, Y, Satoh, T, Yagi, H, Yanaka, S, Yamaguchi, T, Isoda, Y, Iida, S, Okamoto, Y, Kato, K. | | Deposit date: | 2017-05-01 | | Release date: | 2017-11-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conformational effects of N-glycan core fucosylation of immunoglobulin G Fc region on its interaction with Fc gamma receptor IIIa.
Sci Rep, 7, 2017
|
|
5XJF
 
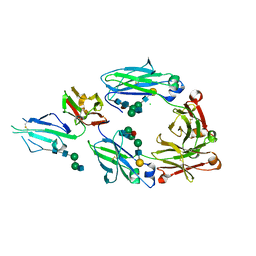 | | Crystal structure of fucosylated IgG Fc Y296W mutant complexed with bis-glycosylated soluble form of Fc gamma receptor IIIa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Sakae, Y, Satoh, T, Yagi, H, Yanaka, S, Yamaguchi, T, Isoda, Y, Iida, S, Okamoto, Y, Kato, K. | | Deposit date: | 2017-05-01 | | Release date: | 2017-11-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Conformational effects of N-glycan core fucosylation of immunoglobulin G Fc region on its interaction with Fc gamma receptor IIIa.
Sci Rep, 7, 2017
|
|
5H5A
 
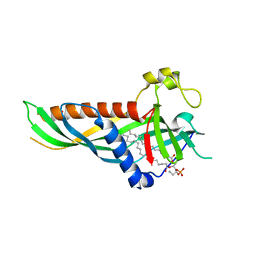 | | Mdm12 from K. lactis (1-239), Lys residues are uniformly dimethyl modified | | Descriptor: | Mitochondrial distribution and morphology protein 12, POTASSIUM ION, [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate | | Authors: | Kawano, S, Quinbara, S, Endo, T. | | Deposit date: | 2016-11-04 | | Release date: | 2017-11-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structure-function insights into direct lipid transfer between membranes by Mmm1-Mdm12 of ERMES
J. Cell Biol., 217, 2018
|
|
5H54
 
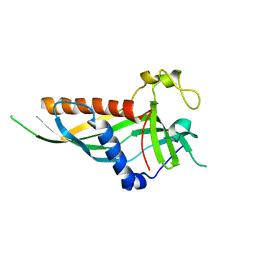 | | Mdm12 from K. lactis 1-239 | | Descriptor: | Mitochondrial distribution and morphology protein 12 | | Authors: | Kawano, S, Quinbara, S, Endo, T. | | Deposit date: | 2016-11-04 | | Release date: | 2017-11-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure-function insights into direct lipid transfer between membranes by Mmm1-Mdm12 of ERMES
J. Cell Biol., 217, 2018
|
|
5H55
 
 | | Mdm12 from K. lactis | | Descriptor: | Mitochondrial distribution and morphology protein 12 | | Authors: | Kawano, S, Quinbara, S. | | Deposit date: | 2016-11-04 | | Release date: | 2017-11-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure-function insights into direct lipid transfer between membranes by Mmm1-Mdm12 of ERMES
J. Cell Biol., 217, 2018
|
|
5H5C
 
 | |
