2JWT
 
 | | Solution structure of Engrailed homeodomain WT | | Descriptor: | Segmentation polarity homeobox protein engrailed | | Authors: | Religa, T.L. | | Deposit date: | 2007-10-24 | | Release date: | 2008-04-01 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Comparison of multiple crystal structures with NMR data for engrailed homeodomain
J.Biomol.Nmr, 40, 2008
|
|
1ZTR
 
 | | Solution structure of Engrailed homeodomain L16A mutant | | Descriptor: | Segmentation polarity homeobox protein engrailed | | Authors: | Religa, T.L, Markson, J.S, Mayor, U, Freund, S.M.V, Fersht, A.R. | | Deposit date: | 2005-05-27 | | Release date: | 2005-10-18 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a protein denatured state and folding intermediate.
Nature, 437, 2005
|
|
2KU1
 
 | |
2P81
 
 | | Engrailed homeodomain helix-turn-helix motif | | Descriptor: | Segmentation polarity homeobox protein engrailed | | Authors: | Religa, T.L. | | Deposit date: | 2007-03-21 | | Release date: | 2007-06-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The helix-turn-helix motif as an ultrafast independently folding domain: The pathway of folding of Engrailed homeodomain.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2KU2
 
 | |
2V0E
 
 | |
2V0F
 
 | |
2LKS
 
 | | Ff11-60 | | Descriptor: | Pre-mRNA-processing factor 40 homolog A | | Authors: | Barette, J, Velyvis, A, Religa, T.L, Korzhnev, D.M, Kay, L.E. | | Deposit date: | 2011-10-19 | | Release date: | 2012-01-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Cross-Validation of the Structure of a Transiently Formed and Low Populated FF Domain Folding Intermediate Determined by Relaxation Dispersion NMR and CS-Rosetta.
J.Phys.Chem.B, 116, 2012
|
|
2KZG
 
 | | A Transient and Low Populated Protein Folding Intermediate at Atomic Resolution | | Descriptor: | Pre-mRNA-processing factor 40 homolog A | | Authors: | Korzhnev, D.M, Religa, T.L, Banachewicz, W, Fersht, A.R, Kay, L.E. | | Deposit date: | 2010-06-17 | | Release date: | 2010-09-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A transient and low-populated protein-folding intermediate at atomic resolution.
Science, 329, 2010
|
|
2L9V
 
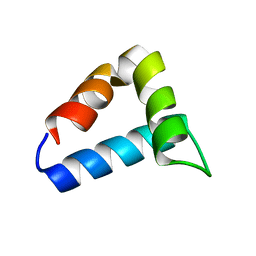 | | NMR structure of the FF domain L24A mutant's folding transition state | | Descriptor: | Pre-mRNA-processing factor 40 homolog A | | Authors: | Korzhnev, D.M, Vernon, R.M, Religa, T.L, Hansen, A, Baker, D, Fersht, A.R, Kay, L.E. | | Deposit date: | 2011-02-24 | | Release date: | 2011-09-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Nonnative interactions in the FF domain folding pathway from an atomic resolution structure of a sparsely populated intermediate: an NMR relaxation dispersion study.
J.Am.Chem.Soc., 133, 2011
|
|
1SS1
 
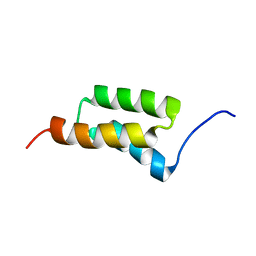 | | STAPHYLOCOCCAL PROTEIN A, B-DOMAIN, Y15W MUTANT, NMR, 25 STRUCTURES | | Descriptor: | Immunoglobulin G binding protein A | | Authors: | Sato, S, Religa, T.L, Daggett, V, Fersht, A.R. | | Deposit date: | 2004-03-23 | | Release date: | 2004-04-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | From The Cover: Testing protein-folding simulations by experiment: B domain of protein A.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
