2JHQ
 
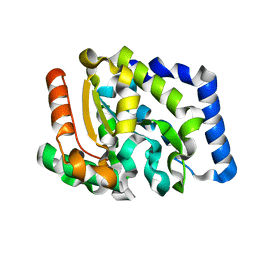 | | Crystal structure of Uracil DNA-glycosylase from Vibrio cholerae | | Descriptor: | CHLORIDE ION, URACIL DNA-GLYCOSYLASE | | Authors: | Raeder, I.L.U, Moe, E, Willassen, N.P, Smalas, A.O, Leiros, I. | | Deposit date: | 2007-02-23 | | Release date: | 2008-06-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of Uracil-DNA N-Glycosylase (Ung) from Vibrio Cholerae. Mapping Temperature Adaptation Through Structural and Mutational Analysis.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
5AFD
 
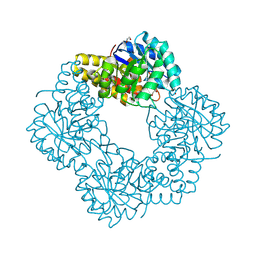 | | Native structure of N-acetylneuramininate lyase (sialic acid aldolase) from Aliivibrio salmonicida | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, N-ACETYLNEURAMINATE LYASE | | Authors: | Gurung, M.K, Altermark, B, Rader, I.L.U, Helland, R, Smalas, A.O. | | Deposit date: | 2015-01-21 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Features and structure of a cold active N-acetylneuraminate lyase.
Plos One, 14, 2019
|
|
6F04
 
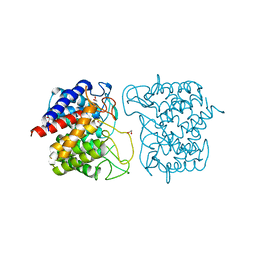 | | N-acetylglucosamine-2-epimerase | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, N-acetylglucosamine-2-epimerase | | Authors: | Halsoer, M.J, Rothweiler, U. | | Deposit date: | 2017-11-17 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | The crystal structure of the N-acetylglucosamine 2-epimerase from Nostoc sp. KVJ10 reveals the true dimer.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
