7VN8
 
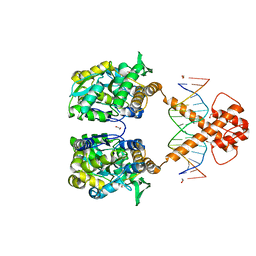 | | Crystal structure of MBP-fused BIL1/BZR1 (21-90) in complex with double-stranded DNA contaning GTCACGTGAC | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*TP*TP*GP*TP*CP*AP*CP*GP*TP*GP*AP*CP*AP*AP*A)-3'), Maltodextrin-binding protein,Protein BRASSINAZOLE-RESISTANT 1, ... | | Authors: | Nosaki, S, Tanokura, M, Miyakawa, T. | | Deposit date: | 2021-10-10 | | Release date: | 2022-12-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Brassinosteroid-induced gene repression requires specific and tight promoter binding of BIL1/BZR1 via DNA shape readout.
Nat.Plants, 8, 2022
|
|
7VN6
 
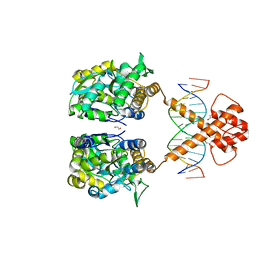 | | Crystal structure of MBP-fused BIL1/BZR1 (21-90) in complex with double-stranded DNA contaning CGCACGTGCG | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*TP*TP*CP*GP*CP*AP*CP*GP*TP*GP*CP*GP*AP*AP*A)-3'), Maltodextrin-binding protein,Protein BRASSINAZOLE-RESISTANT 1, ... | | Authors: | Nosaki, S, Tanokura, M, Miyakawa, T. | | Deposit date: | 2021-10-10 | | Release date: | 2022-12-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Brassinosteroid-induced gene repression requires specific and tight promoter binding of BIL1/BZR1 via DNA shape readout.
Nat.Plants, 8, 2022
|
|
7VN7
 
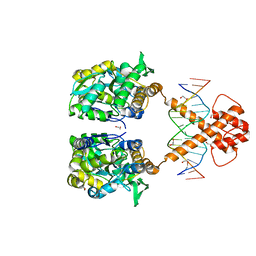 | | Crystal structure of MBP-fused BIL1/BZR1 (21-90) in complex with double-stranded DNA contaning GACACGTGTC | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*TP*TP*GP*AP*CP*AP*CP*GP*TP*GP*TP*CP*AP*AP*A)-3'), Maltodextrin-binding protein,Protein BRASSINAZOLE-RESISTANT 1, ... | | Authors: | Nosaki, S, Tanokura, M, Miyakawa, T. | | Deposit date: | 2021-10-10 | | Release date: | 2022-12-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Brassinosteroid-induced gene repression requires specific and tight promoter binding of BIL1/BZR1 via DNA shape readout.
Nat.Plants, 8, 2022
|
|
7VN5
 
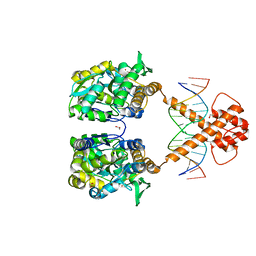 | | Crystal structure of MBP-fused BIL1/BZR1 (21-90) in complex with double-stranded DNA contaning TTCACGTGAA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*TP*TP*TP*TP*CP*AP*CP*GP*TP*GP*AP*AP*AP*AP*A)-3'), Maltodextrin-binding protein,Protein BRASSINAZOLE-RESISTANT 1, ... | | Authors: | Nosaki, S, Tanokura, M, Miyakawa, T. | | Deposit date: | 2021-10-10 | | Release date: | 2022-12-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Brassinosteroid-induced gene repression requires specific and tight promoter binding of BIL1/BZR1 via DNA shape readout.
Nat.Plants, 8, 2022
|
|
7VN3
 
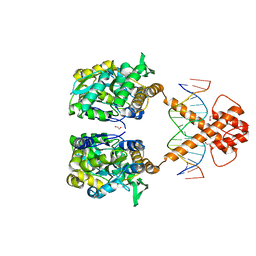 | | Crystal structure of MBP-fused BIL1/BZR1 (21-90) in complex with double-stranded DNA contaning CACACGTGTG | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*TP*TP*CP*AP*CP*AP*CP*GP*TP*GP*TP*GP*AP*AP*A)-3'), Maltodextrin-binding protein,Protein BRASSINAZOLE-RESISTANT 1, ... | | Authors: | Nosaki, S, Tanokura, M, Miyakawa, T. | | Deposit date: | 2021-10-10 | | Release date: | 2022-12-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Brassinosteroid-induced gene repression requires specific and tight promoter binding of BIL1/BZR1 via DNA shape readout.
Nat.Plants, 8, 2022
|
|
7VN4
 
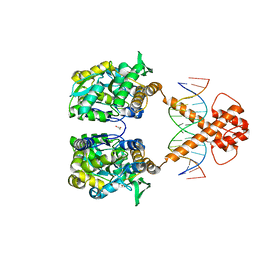 | | Crystal structure of MBP-fused BIL1/BZR1 (21-90) in complex with double-stranded DNA contaning TCCACGTGGA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*TP*TP*TP*CP*CP*AP*CP*GP*TP*GP*AP*AP*AP*AP*A)-3'), Maltodextrin-binding protein,Protein BRASSINAZOLE-RESISTANT 1, ... | | Authors: | Nosaki, S, Tanokura, M, Miyakawa, T. | | Deposit date: | 2021-10-10 | | Release date: | 2022-12-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Brassinosteroid-induced gene repression requires specific and tight promoter binding of BIL1/BZR1 via DNA shape readout.
Nat.Plants, 8, 2022
|
|
7VN2
 
 | | Crystal structure of MBP-fused BIL1/BZR1 (21-90) in complex with double-stranded DNA contaning ATCACGTGAT | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*TP*TP*AP*TP*CP*AP*CP*GP*TP*GP*AP*TP*AP*AP*A)-3'), Maltodextrin-binding protein,Protein BRASSINAZOLE-RESISTANT 1, ... | | Authors: | Nosaki, S, Tanokura, M, Miyakawa, T. | | Deposit date: | 2021-10-10 | | Release date: | 2022-12-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Brassinosteroid-induced gene repression requires specific and tight promoter binding of BIL1/BZR1 via DNA shape readout.
Nat.Plants, 8, 2022
|
|
5ZD4
 
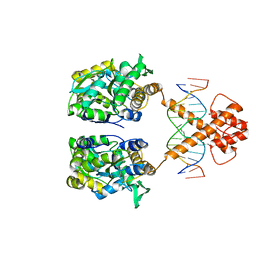 | | Crystal structure of MBP-fused BIL1/BZR1 in complex with double-stranded DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*TP*TP*CP*AP*CP*AP*CP*GP*TP*GP*TP*GP*AP*AP*A)-3'), Maltose-binding periplasmic protein,Protein BRASSINAZOLE-RESISTANT 1, ... | | Authors: | Nosaki, S, Miyakawa, T, Xu, Y, Nakamura, A, Hirabayashi, K, Tanokura, M. | | Deposit date: | 2018-02-22 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structural basis for brassinosteroid response by BIL1/BZR1.
Nat Plants, 4, 2018
|
|
1GEE
 
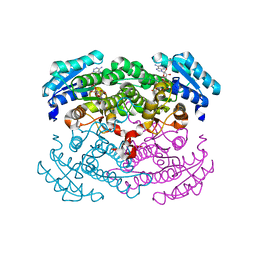 | | Crystal structure of glucose dehydrogenase mutant Q252L complexed with NAD+ | | Descriptor: | GLUCOSE 1-DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Yamamoto, K, Kurisu, G, Kusunoki, M, Tabata, S, Urabe, I, Osaki, S. | | Deposit date: | 2000-11-07 | | Release date: | 2003-08-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural analysis of stability-increasing mutants of glucose dehydrogenase
To be Published
|
|
1G6K
 
 | | Crystal structure of glucose dehydrogenase mutant E96A complexed with NAD+ | | Descriptor: | GLUCOSE 1-DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Yamamoto, K, Kurisu, G, Kusunoki, M, Tabata, S, Urabe, I, Osaki, S. | | Deposit date: | 2000-11-06 | | Release date: | 2003-08-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis of stability-increasing mutants of glucose dehydrogenase
To be Published
|
|
3AJ7
 
 | | Crystal Structure of isomaltase from Saccharomyces cerevisiae | | Descriptor: | CALCIUM ION, Oligo-1,6-glucosidase | | Authors: | Yamamoto, K, Miyake, H, Kusunoki, M, Osaki, S. | | Deposit date: | 2010-05-26 | | Release date: | 2010-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structures of isomaltase from Saccharomyces cerevisiae and in complex with its competitive inhibitor maltose
Febs J., 277, 2010
|
|
1GCO
 
 | | CRYSTAL STRUCTURE OF GLUCOSE DEHYDROGENASE COMPLEXED WITH NAD+ | | Descriptor: | GLUCOSE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Yamamoto, K, Kurisu, G, Kusunoki, M, Tabata, S, Urabe, I, Osaki, S. | | Deposit date: | 2000-08-07 | | Release date: | 2001-02-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of glucose dehydrogenase from Bacillus megaterium IWG3 at 1.7 A resolution.
J.Biochem., 129, 2001
|
|
3AXH
 
 | | Crystal structure of isomaltase in complex with isomaltose | | Descriptor: | CALCIUM ION, Oligo-1,6-glucosidase IMA1, alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose | | Authors: | Yamamoto, K, Miyake, H, Kusunoki, M, Osaki, S. | | Deposit date: | 2011-04-06 | | Release date: | 2011-10-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Steric hindrance by 2 amino acid residues determines the substrate specificity of isomaltase from Saccharomyces cerevisiae
J.Biosci.Bioeng., 112, 2011
|
|
3AXI
 
 | | Crystal structure of isomaltase in complex with maltose | | Descriptor: | CALCIUM ION, Oligo-1,6-glucosidase IMA1, alpha-D-glucopyranose | | Authors: | Yamamoto, K, Miyake, H, Kusunoki, M, Osaki, S. | | Deposit date: | 2011-04-06 | | Release date: | 2011-10-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Steric hindrance by 2 amino acid residues determines the substrate specificity of isomaltase from Saccharomyces cerevisiae
J.Biosci.Bioeng., 112, 2011
|
|
3A4A
 
 | | Crystal structure of isomaltase from Saccharomyces cerevisiae | | Descriptor: | CALCIUM ION, Oligo-1,6-glucosidase, alpha-D-glucopyranose | | Authors: | Yamamoto, K, Miyake, H, Kusunoki, M, Osaki, S. | | Deposit date: | 2009-07-01 | | Release date: | 2010-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of isomaltase from Saccharomyces cerevisiae and in complex with its competitive inhibitor maltose
Febs J., 277, 2010
|
|
3A47
 
 | |
5XFM
 
 | | Crystal structure of beta-arabinopyranosidase | | Descriptor: | Alpha-glucosidase, CALCIUM ION | | Authors: | Kato, K, Okuyama, M, Yao, M. | | Deposit date: | 2017-04-10 | | Release date: | 2018-02-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A novel glycoside hydrolase family 97 enzyme: Bifunctional beta-l-arabinopyranosidase/ alpha-galactosidase from Bacteroides thetaiotaomicron.
Biochimie, 142, 2017
|
|
5Z7Y
 
 | | Crystal structure of Striga hermonthica HTL7 (ShHTL7) | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, Hyposensitive to light 7, ... | | Authors: | Xu, Y, Miyakawa, T, Nakamura, A, Tanokura, M. | | Deposit date: | 2018-01-30 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of HTL and D14 proteins reveals the basis for ligand selectivity in Striga.
Nat Commun, 9, 2018
|
|
5Z7Z
 
 | | Crystal structure of Striga hermonthica Dwarf14 (ShD14) | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Dwarf 14, ... | | Authors: | Xu, Y, Miyakawa, T, Nakamura, A, Tanokura, M. | | Deposit date: | 2018-01-30 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.978 Å) | | Cite: | Structural analysis of HTL and D14 proteins reveals the basis for ligand selectivity in Striga.
Nat Commun, 9, 2018
|
|
5Z7W
 
 | | Crystal structure of Striga hermonthica HTL1 (ShHTL1) | | Descriptor: | GLYCEROL, Hyposensitive to light 1, MAGNESIUM ION, ... | | Authors: | Xu, Y, Miyakawa, T, Nakamura, A, Tanokura, M. | | Deposit date: | 2018-01-30 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.657 Å) | | Cite: | Structural analysis of HTL and D14 proteins reveals the basis for ligand selectivity in Striga.
Nat Commun, 9, 2018
|
|
5Z7X
 
 | | Crystal structure of Striga hermonthica HTL4 (ShHTL4) | | Descriptor: | 1,2-ETHANEDIOL, Hyposensitive to light 4, MAGNESIUM ION | | Authors: | Xu, Y, Miyakawa, T, Nakamura, A, Tanokura, M. | | Deposit date: | 2018-01-30 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.055 Å) | | Cite: | Structural analysis of HTL and D14 proteins reveals the basis for ligand selectivity in Striga.
Nat Commun, 9, 2018
|
|
